7W93
 
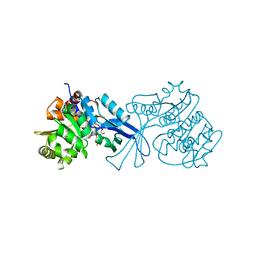 | | Crystal structure of E.coli pseudouridine kinase PsuK complexed with N1-methyl-pseudouridine | | Descriptor: | 5-[(2S,3R,4S,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)oxolan-2-yl]-1-methyl-pyrimidine-2,4-dione, POTASSIUM ION, PfkB domain protein | | Authors: | Li, K.J, Li, X.J, Wu, B.X. | | Deposit date: | 2021-12-09 | | Release date: | 2022-06-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure Characterization of Escherichia coli Pseudouridine Kinase PsuK.
Front Microbiol, 13, 2022
|
|
1BBA
 
 | |
7VU7
 
 | |
6KX9
 
 | |
7CYB
 
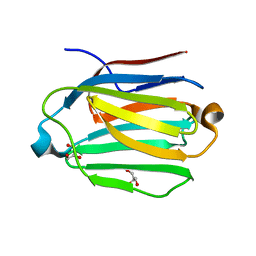 | | Saimiri boliviensis boliviensis galectin-13 with glycerol | | Descriptor: | GLYCEROL, Galectin | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
7CYA
 
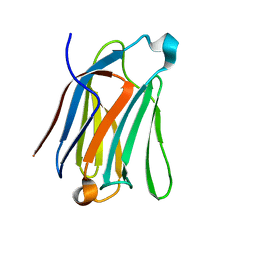 | | Saimiri boliviensis boliviensis galectin-13 with lactose | | Descriptor: | Galectin, beta-D-galactopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Su, J. | | Deposit date: | 2020-09-03 | | Release date: | 2021-06-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Actin binding to galectin-13/placental protein-13 occurs independently of the galectin canonical ligand-binding site.
Glycobiology, 31, 2021
|
|
8JQE
 
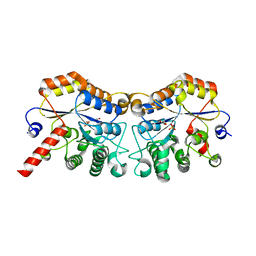 | | Structure of CmCBDA in complex with Mn2+ and glycerol | | Descriptor: | GLYCEROL, MANGANESE (II) ION, PHOSPHATE ION, ... | | Authors: | Li, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Insights into the Catalytic Activity of Cyclobacterium marinum N -Acetylglucosamine Deacetylase.
J.Agric.Food Chem., 72, 2024
|
|
8JQF
 
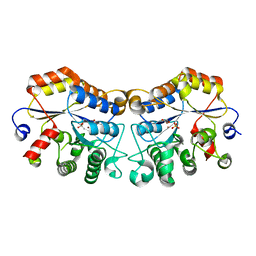 | | Structure of CmCBDA in complex with Ni2+ and Glycerol | | Descriptor: | GLYCEROL, NICKEL (II) ION, SULFATE ION, ... | | Authors: | Li, X. | | Deposit date: | 2023-06-14 | | Release date: | 2024-01-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights into the Catalytic Activity of Cyclobacterium marinum N -Acetylglucosamine Deacetylase.
J.Agric.Food Chem., 72, 2024
|
|
7VS7
 
 | | Crystal structure of the ectodomain of OsCERK1 in complex with chitin hexamer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitin elicitor receptor kinase 1, ... | | Authors: | Li, X. | | Deposit date: | 2021-10-26 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Structural insight into chitin perception by chitin elicitor receptor kinase 1 of Oryza sativa.
J Integr Plant Biol, 65, 2023
|
|
3DOY
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3i | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-chloro-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DP3
 
 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3q | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-tert-butyl-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
7YV8
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with golden hamster ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme, Spike glycoprotein, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.94 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
7YVU
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.2 RBD in complex with mouse ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike protein S1, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Chai, Y, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-08-19 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
8SDG
 
 | |
7YW2
 
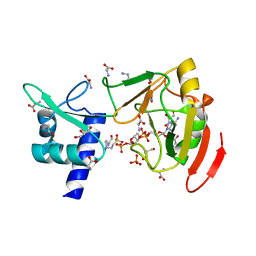 | | Crystal structure of tRNA 2'-phosphotransferase from Mus musculus | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
7YW3
 
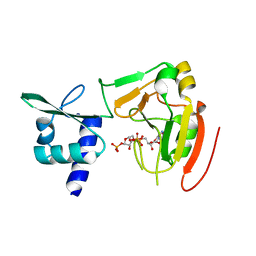 | | Crystal structure of tRNA 2'-phosphotransferase from Homo sapiens | | Descriptor: | 1,2-ETHANEDIOL, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-3,4-bis(oxidanyl)-5-phosphonooxy-oxolan-2-yl]methyl hydrogen phosphate, tRNA 2'-phosphotransferase 1 | | Authors: | Yang, X.Y, Liu, X.H. | | Deposit date: | 2022-08-21 | | Release date: | 2023-07-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical insights into the molecular mechanism of TRPT1 for nucleic acid ADP-ribosylation.
Nucleic Acids Res., 51, 2023
|
|
7YW4
 
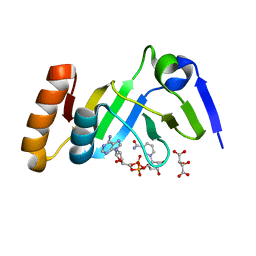 | |
8SDF
 
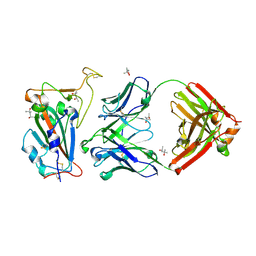 | |
8R7Q
 
 | |
8R7P
 
 | |
8SDH
 
 | |
3LRC
 
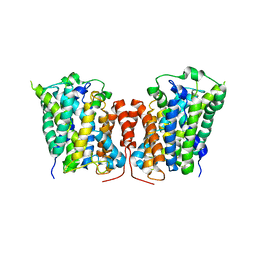 | | Structure of E. coli AdiC (P1) | | Descriptor: | Arginine/agmatine antiporter | | Authors: | Gao, X, Lu, F, Zhou, L, Shi, Y. | | Deposit date: | 2010-02-11 | | Release date: | 2010-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.004 Å) | | Cite: | Structure and mechanism of an amino acid antiporter
Science, 324, 2009
|
|
4KFW
 
 | |
4KFV
 
 | |
8H06
 
 | | Cryo-EM structure of SARS-CoV-2 Omicron BA.4/5 RBD in complex with human ACE2 (local refinement) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhao, Z.N, Xie, Y.F, Qi, J.X, Gao, G.F. | | Deposit date: | 2022-09-28 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis for receptor binding and broader interspecies receptor recognition of currently circulating Omicron sub-variants.
Nat Commun, 14, 2023
|
|
