8OTC
 
 | |
8OTF
 
 | |
8OTE
 
 | |
8OT6
 
 | |
8OTJ
 
 | |
8OT9
 
 | |
5B1M
 
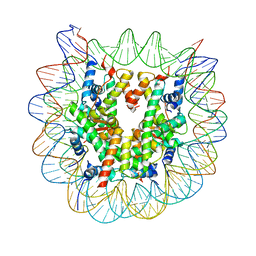 | | The mouse nucleosome structure containing H3.1 | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B type 3-A, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
5B1L
 
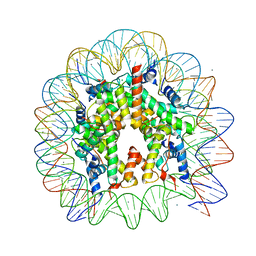 | | The mouse nucleosome structure containing H3t | | Descriptor: | CHLORIDE ION, DNA (146-MER), Histone H2A type 1, ... | | Authors: | Urahama, T, Machida, S, Horikoshi, N, Osakabe, A, Tachiwana, H, Taguchi, H, Kurumizaka, H. | | Deposit date: | 2015-12-08 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Testis-Specific Histone Variant H3t Gene Is Essential for Entry into Spermatogenesis
Cell Rep, 18, 2017
|
|
7DB0
 
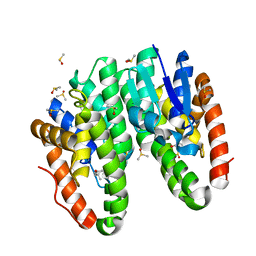 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in dimedone-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB1
 
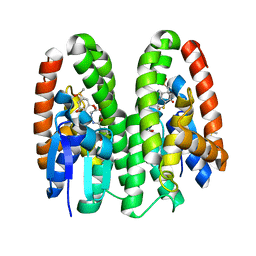 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in dimedone- and glutathione-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAZ
 
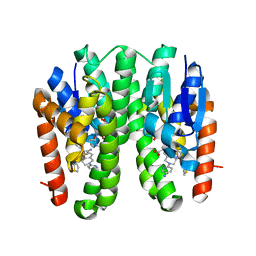 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP015- and GSH-bound form | | Descriptor: | 6-(6,7-dihydro-4H-thieno[3,2-c]pyridin-5-yl)pyridin-3-amine, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB2
 
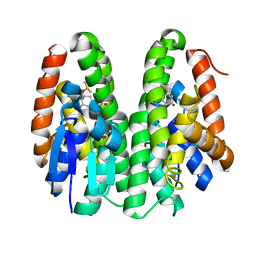 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in GS-dimedone-bound form | | Descriptor: | 5,5-DIMETHYLCYCLOHEXANE-1,3-DIONE, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAY
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP013-, and GSH-bound form | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-[(5-tert-butyl-2-methyl-phenyl)sulfonylamino]benzoic acid, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB4
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP012- and glutathione-bound form | | Descriptor: | 1-[(4-fluorophenyl)methyl]-2,2-bis(oxidanylidene)thieno[3,2-c][1,2]thiazin-4-one, GLUTATHIONE, Glutathione S-transferase E14 | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DAX
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP013-bound form | | Descriptor: | 2-[(5-tert-butyl-2-methyl-phenyl)sulfonylamino]benzoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7000097 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
7DB3
 
 | | Crystal structure of Drosophila melanogaster Noppera-bo, glutathione S-transferase epsilon 14 (DmGSTE14), in TDP011-bound form | | Descriptor: | 4-bromanyl-2-[[2-[(E)-1-(3-methoxyphenyl)ethylideneamino]propan-2-ylamino]methyl]phenol, DIMETHYL SULFOXIDE, GLUTATHIONE, ... | | Authors: | Koiwai, K, Inaba, K, Yumoto, F, Senda, T, Niwa, R. | | Deposit date: | 2020-10-18 | | Release date: | 2021-02-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Non-steroidal inhibitors of Drosophila melanogaster steroidogenic glutathione S -transferase Noppera-bo
J Pestic Sci, 46, 2021
|
|
3VYD
 
 | | Human renin in complex with inhibitor 6 | | Descriptor: | (3S,5R)-5-{[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]methyl}-N-(3-methylbutyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, Renin | | Authors: | Takahashi, M, Hanzawa, H. | | Deposit date: | 2012-09-24 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Design and discovery of new (3S,5R)-5-[4-(2-chlorophenyl)-2,2-dimethyl-5-oxopiperazin-1-yl]piperidine-3-carboxamides as potent renin inhibitors
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6OYH
 
 | | Crystal structure of MraY bound to carbacaprazamycin | | Descriptor: | (5S)-5'-O-(5-amino-5-deoxy-beta-D-ribofuranosyl)-5'-C-[(2S,5S,6S)-5-carboxy-6-heptadecyl-1,4-dimethyl-3-oxo-1,4-diazepan-2-yl]uridine, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-14 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
6OYZ
 
 | | Crystal structure of MraY bound to capuramycin | | Descriptor: | (2~{S},3~{S},4~{S})-2-[(1~{R})-2-azanyl-1-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3-methoxy-4-oxidanyl-oxolan-2-yl]-2-oxidanylidene-ethoxy]-3,4-bis(oxidanyl)-~{N}-[(3~{S})-2-oxidanylideneazepan-3-yl]-3,4-dihydro-2~{H}-pyran-6-carboxamide, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
6OZ6
 
 | | Crystal structure of MraY bound to 3'-hydroxymureidomycin A | | Descriptor: | (2~{S})-2-[[(2~{S})-1-[[(2~{S},3~{S})-3-[[(2~{S})-2-azanyl-3-(3-hydroxyphenyl)propanoyl]-methyl-amino]-1-[[(~{Z})-[(3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-ylidene]methyl]amino]-1-oxidanylidene-butan-2-yl]amino]-4-methylsulfanyl-1-oxidanylidene-butan-2-yl]carbamoylamino]-3-(3-hydroxyphenyl)propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2019-05-15 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Chemical logic of MraY inhibition by antibacterial nucleoside natural products.
Nat Commun, 10, 2019
|
|
4TWL
 
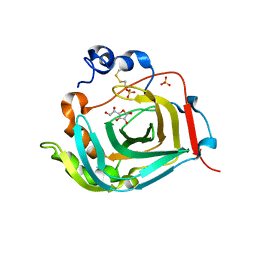 | | Crystal structure of dioscorin complexed with ascorbate | | Descriptor: | ASCORBIC ACID, Dioscorin 5, SULFATE ION | | Authors: | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
4TWM
 
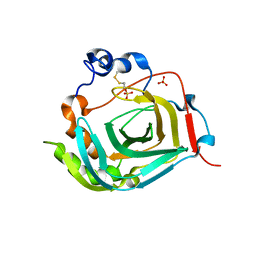 | | Crystal structure of dioscorin from Dioscorea japonica | | Descriptor: | Dioscorin 5, SULFATE ION | | Authors: | Xue, Y.L, Miyakawa, T, Nakamura, A, Tanokura, M. | | Deposit date: | 2014-07-01 | | Release date: | 2015-04-01 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Yam Tuber Storage Protein Reduces Plant Oxidants Using the Coupled Reactions as Carbonic Anhydrase and Dehydroascorbate Reductase
Mol Plant, 8, 2015
|
|
6ILI
 
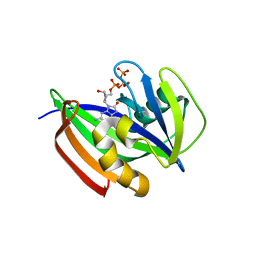 | | Crystal structure of human MTH1(G2K/D120N mutant) in complex with 8-oxo-dGTP at pH 6.5 | | Descriptor: | 7,8-dihydro-8-oxoguanine triphosphatase, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE | | Authors: | Nakamura, T, Waz, S, Hirata, K, Nakabeppu, Y, Yamagata, Y. | | Deposit date: | 2018-10-18 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Kinetic Studies of the Human Nudix Hydrolase MTH1 Reveal the Mechanism for Its Broad Substrate Specificity
J. Biol. Chem., 292, 2017
|
|
1B5Z
 
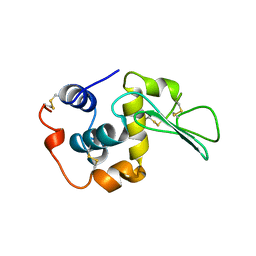 | | CONTRIBUTION OF HYDROGEN BONDS TO THE CONFORMATIONAL STABILITY OF HUMAN LYSOZYME: CALORIMETRY AND X-RAY ANALYSIS OF SIX SER->ALA MUTANTS | | Descriptor: | LYSOZYME | | Authors: | Takano, K, Yamagata, Y, Kubota, M, Funahashi, J, Fujii, S, Yutani, K. | | Deposit date: | 1999-01-11 | | Release date: | 1999-02-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Contribution of hydrogen bonds to the conformational stability of human lysozyme: calorimetry and X-ray analysis of six Ser --> Ala mutants.
Biochemistry, 38, 1999
|
|
5H72
 
 | | Structure of the periplasmic domain of FliP | | Descriptor: | Flagellar biosynthetic protein FliP | | Authors: | Fukumura, T, Kawaguchi, T, Saijo-Hamano, Y, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-11-16 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Assembly and stoichiometry of the core structure of the bacterial flagellar type III export gate complex
PLoS Biol., 15, 2017
|
|
