6LBE
 
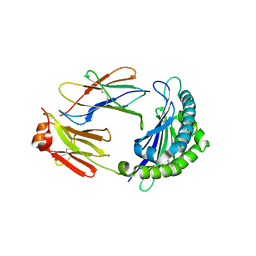 | |
6E4R
 
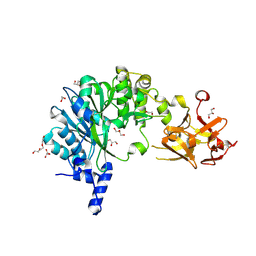 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9B | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.061 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
6E4Q
 
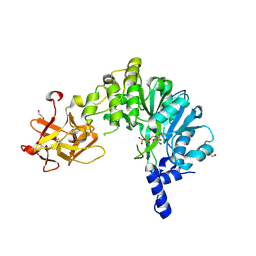 | | Crystal Structure of the Drosophila Melanogaster Polypeptide N-Acetylgalactosaminyl Transferase PGANT9A in Complex with UDP and Mn2+ | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Samara, N.L, Tabak, L.A, Ten Hagen, K.G. | | Deposit date: | 2018-07-18 | | Release date: | 2018-09-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | A molecular switch orchestrates enzyme specificity and secretory granule morphology.
Nat Commun, 9, 2018
|
|
4WQ6
 
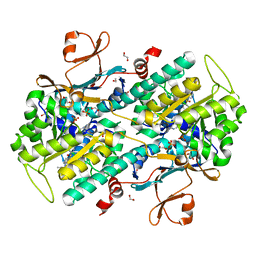 | | The crystal structure of human Nicotinamide phosphoribosyltransferase (NAMPT) in complex with N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide inhibitor (compound 21) | | Descriptor: | 1,2-ETHANEDIOL, N-(4-{(S)-[1-(2-methylpropyl)piperidin-4-yl]sulfinyl}benzyl)furo[2,3-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, ... | | Authors: | Li, D, Wang, W. | | Deposit date: | 2014-10-21 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Identification of nicotinamide phosphoribosyltransferase (NAMPT) inhibitors with no evidence of CYP3A4 time-dependent inhibition and improved aqueous solubility.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6MC1
 
 | | Structure of MAP kinase phosphatase 5 in complex with 3,3-dimethyl-1-((9-(methylthio)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl)thio)butan-2-one, an allosteric inhibitor | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 3,3-dimethyl-1-{[9-(methylsulfanyl)-5,6-dihydrothieno[3,4-h]quinazolin-2-yl]sulfanyl}butan-2-one, ACETATE ION, ... | | Authors: | Gannam, Z.T.K, Anderson, K.S, Bennett, A.M, Lolis, E. | | Deposit date: | 2018-08-30 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | An allosteric site on MKP5 reveals a strategy for small-molecule inhibition.
Sci.Signal., 13, 2020
|
|
4ZFC
 
 | | Crystal structure of AKR1C3 complexed with glicazide | | Descriptor: | Aldo-keto reductase family 1 member C3, N-[(3aR,6aS)-hexahydrocyclopenta[c]pyrrol-2(1H)-ylcarbamoyl]-4-methylbenzenesulfonamide, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhao, Y, Zhrng, X, Hu, X. | | Deposit date: | 2015-04-21 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vitro inhibition of AKR1Cs by sulphonylureas and the structural basis
Chem.Biol.Interact., 240, 2015
|
|
7AXU
 
 | |
7AXQ
 
 | |
7AXP
 
 | |
7AXX
 
 | |
7AXS
 
 | |
4YJN
 
 | |
5XMM
 
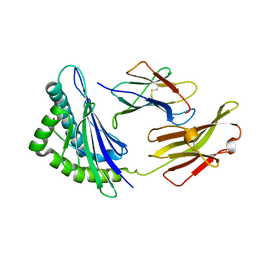 | | FLA-E*01801-167W/S | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
5ZZD
 
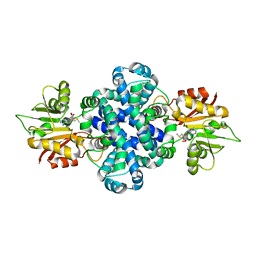 | | Crystal structure of a protein from Aspergillus flavus | | Descriptor: | O-methyltransferase lepI, S-ADENOSYLMETHIONINE | | Authors: | Chang, Z.Y, Liu, W.D, Chen, C.C, Guo, R.T. | | Deposit date: | 2018-05-31 | | Release date: | 2019-06-05 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of LepI, a multifunctional SAM-dependent enzyme which catalyzes pericyclic reactions in leporin biosynthesis.
Org.Biomol.Chem., 17, 2019
|
|
5XMF
 
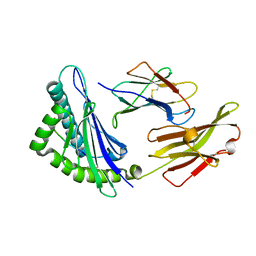 | | Crystal structure of feline MHC class I for 2,1 angstrom | | Descriptor: | Beta-2-microglobulin, Gag polyprotein, MHC class I antigen alpha chain | | Authors: | Liang, R, Sun, Y, Wang, J, Wu, Y, Zhang, N, Xia, C. | | Deposit date: | 2017-05-15 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Major Histocompatibility Complex Class I (FLA-E*01801) Molecular Structure in Domestic Cats Demonstrates Species-Specific Characteristics in Presenting Viral Antigen Peptides
J. Virol., 92, 2018
|
|
8KHD
 
 | | The interface structure of Omicron RBD binding to 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817, Light chain of 5817, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
8KHC
 
 | | SARS-CoV-2 Omicron spike in complex with 5817 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of 5817 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2023-08-21 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Identification of a broad sarbecovirus neutralizing antibody targeting a conserved epitope on the receptor-binding domain.
Cell Rep, 43, 2024
|
|
5J8V
 
 | |
6KZD
 
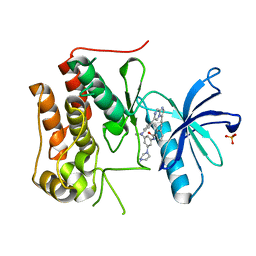 | | Crystal structure of TRKc in complex with 3-((6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl)ethynyl)- N-(3-isopropyl-5-((4-methylpiperazin-1-yl)methyl)phenyl)-2- methylbenzamide | | Descriptor: | 3-[2-[6-(4-aminophenyl)imidazo[1,2-a]pyrazin-3-yl]ethynyl]-2-methyl-~{N}-[3-(4-methylpiperazin-1-yl)-5-propan-2-yl-phenyl]benzamide, NT-3 growth factor receptor, PHOSPHATE ION | | Authors: | Wang, Y, Zhang, Z.M. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-09 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.708 Å) | | Cite: | Design, synthesis and biological evaluation of 3-(imidazo[1,2-a]pyrazin-3-ylethynyl)-2-methylbenzamides as potent and selective pan-tropomyosin receptor kinase (TRK) inhibitors.
Eur.J.Med.Chem., 179, 2019
|
|
4Q65
 
 | | Structure of the E. coli Peptide Transporter YbgH | | Descriptor: | Dipeptide permease D | | Authors: | Zhang, C, Zhao, Y, Mao, G, Liu, M, Wang, X. | | Deposit date: | 2014-04-21 | | Release date: | 2014-08-13 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of the E. coli peptide transporter YbgH.
Structure, 22, 2014
|
|
4FZF
 
 | | Crystal structure of MST4-MO25 complex with DKI | | Descriptor: | 5-AMINO-3-{[4-(AMINOSULFONYL)PHENYL]AMINO}-N-(2,6-DIFLUOROPHENYL)-1H-1,2,4-TRIAZOLE-1-CARBOTHIOAMIDE, Calcium-binding protein 39, Serine/threonine-protein kinase MST4 | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4FZD
 
 | | Crystal structure of MST4-MO25 complex with WSF motif | | Descriptor: | C-terminal peptide from Serine/threonine-protein kinase MST4, Calcium-binding protein 39, GLYCEROL, ... | | Authors: | Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the MST4 in Complex with MO25 Provides Insights into Its Activation Mechanism
Structure, 21, 2013
|
|
4M84
 
 | |
7AT6
 
 | | Structure of thaumatin collected by femtosecond serial crystallography on a COC membrane | | Descriptor: | L(+)-TARTARIC ACID, R-1,2-PROPANEDIOL, SODIUM ION, ... | | Authors: | Martiel, I, Marsh, M, Vera, L, Huang, C.Y, Olieric, V, Leonarski, P, Nass, K, Padeste, C, Karpik, A, Wang, M, Pedrini, B. | | Deposit date: | 2020-10-29 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Commissioning results from the SwissMX instrument for fixed target macromolecular crystallography at SwissFEL
To Be Published
|
|
3V51
 
 | |
