7D3C
 
 | |
6GZM
 
 | | Crystal Structure of Human CKIdelta with A86 | | Descriptor: | CITRIC ACID, Casein kinase I isoform delta, GLYCEROL, ... | | Authors: | Ben-neriah, Y, Venkatachalam, A, Minzel, W, Fink, A, Snir-Alkalay, I, Vacca, J. | | Deposit date: | 2018-07-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Small Molecules Co-targeting CKI alpha and the Transcriptional Kinases CDK7/9 Control AML in Preclinical Models.
Cell, 175, 2018
|
|
6L7E
 
 | |
6L7I
 
 | |
4FLH
 
 | | Crystal structure of human PI3K-gamma in complex with AMG511 | | Descriptor: | 4-(2-[(5-fluoro-6-methoxypyridin-3-yl)amino]-5-{(1R)-1-[4-(methylsulfonyl)piperazin-1-yl]ethyl}pyridin-3-yl)-6-methyl-1,3,5-triazin-2-amine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selective Class I Phosphoinositide 3-Kinase Inhibitors: Optimization of a Series of Pyridyltriazines Leading to the Identification of a Clinical Candidate, AMG 511.
J.Med.Chem., 55, 2012
|
|
3MXF
 
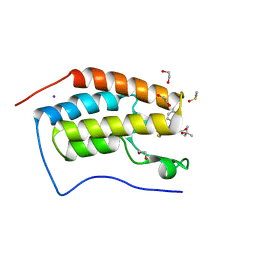 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor JQ1 | | Descriptor: | (6S)-6-(2-tert-butoxy-2-oxoethyl)-4-(4-chlorophenyl)-2,3,9-trimethyl-6,7-dihydrothieno[3,2-f][1,2,4]triazolo[4,3-a][1,4]diazepin-10-ium, 1,2-ETHANEDIOL, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Qi, J, Keates, T, Felletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Bradner, J.E, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-07 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Selective inhibition of BET bromodomains.
Nature, 468, 2010
|
|
7CIR
 
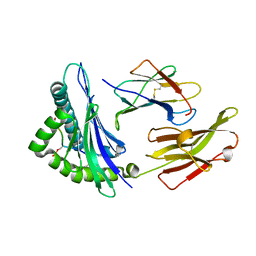 | | Peptide phosphorylation modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIQ
 
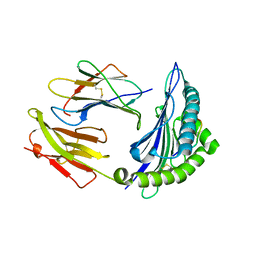 | | Phosphorylation modification of MHC I polypeptide | | Descriptor: | ARG-ARG-PHE-SER-ARG-SER-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
7CIS
 
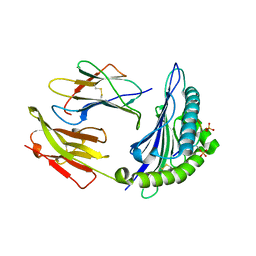 | | Peptide modification of MHC class I molecules | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J, Gao, G.F. | | Deposit date: | 2020-07-08 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
8S2X
 
 | |
8S2V
 
 | |
8S2U
 
 | |
8S2W
 
 | |
6M3G
 
 | | Crystal structure of human HPF1 | | Descriptor: | Histone PARylation factor 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6M3I
 
 | | Crystal structure of HPF1/PARP1 complex | | Descriptor: | BENZAMIDE, Histone PARylation factor 1, Poly [ADP-ribose] polymerase 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6M3H
 
 | | Crystal structure of mouse HPF1 | | Descriptor: | Histone PARylation factor 1 | | Authors: | Sun, F.H, Yun, C.H. | | Deposit date: | 2020-03-03 | | Release date: | 2021-03-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | HPF1 remodels the active site of PARP1 to enable the serine ADP-ribosylation of histones.
Nat Commun, 12, 2021
|
|
6LUO
 
 | | Structure of nurse shark beta-2-microglobulin | | Descriptor: | Beta-2-microglobulin | | Authors: | Xia, C, Wu, Y. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6LUP
 
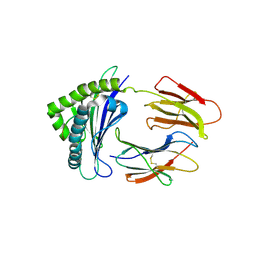 | | Crystal structure of shark MHC CLASS I for 2.3 angstrom | | Descriptor: | Beta-2-microglobulin, MHC class I protein, PHE-ALA-ASN-PHE-PHE-ILE-ARG-GLY-LEU | | Authors: | Wu, Y, Xia, C. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
6N2G
 
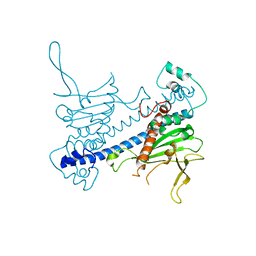 | | Crystal structure of Caenorhabditis elegans NAP1 | | Descriptor: | Nucleosome Assembly Protein | | Authors: | Bhattacharyya, S, DArcy, S. | | Deposit date: | 2018-11-13 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.003 Å) | | Cite: | Characterization of Caenorhabditis elegans Nucleosome Assembly Protein 1 Uncovers the Role of Acidic Tails in Histone Binding.
Biochemistry, 58, 2019
|
|
7DYN
 
 | | Phosphorylation of MHC I peptide | | Descriptor: | ARG-ARG-PHE-SEP-ARG-SEP-PRO-ILE-ARG-ARG, Beta-2-microglobulin, MHC class I antigen | | Authors: | Sun, M.W, Feng, L, Qi, J.X, Liu, W.J. | | Deposit date: | 2021-01-22 | | Release date: | 2022-03-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Phosphosite-dependent presentation of dual phosphorylated peptides by MHC class I molecules.
Iscience, 25, 2022
|
|
4C46
 
 | | ANDREI-N-LVPAS fused to GCN4 adaptors | | Descriptor: | BROMIDE ION, GENERAL CONTROL PROTEIN GCN4 | | Authors: | Albrecht, R, Alva, V, Ammelburg, M, Baer, K, Basina, E, Boichenko, I, Bonhoeffer, F, Braun, V, Chaubey, M, Chauhan, N, Chellamuthu, V.R, Coles, M, Deiss, S, Ewers, C.P, Forouzan, D, Fuchs, A, Groemping, Y, Hartmann, M.D, Hernandez Alvarez, B, Jeganantham, A, Kalev, I, Koenninger, U, Koiwai, K, Kopec, K.O, Korycinski, M, Laudenbach, B, Lehmann, K, Leo, J.C, Linke, D, Marialke, J, Martin, J, Mechelke, M, Michalik, M, Noll, A, Patzer, S.I, Scharfenberg, F, Schueckel, M, Shahid, S.A, Sulz, E, Ursinus, A, Wuertenberger, S, Zhu, H. | | Deposit date: | 2013-08-30 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Your Personalized Protein Structure: Andrei N. Lupas Fused to GCN4 Adaptors.
J.Struct.Biol., 186, 2014
|
|
2Z59
 
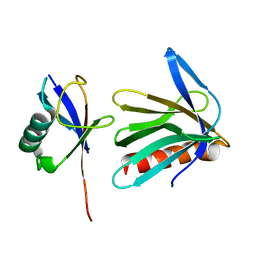 | | Complex Structures of Mouse Rpn13 (22-130aa) and ubiquitin | | Descriptor: | Protein ADRM1, Ubiquitin | | Authors: | Chen, X, Schreiner, P, Groll, M, Walters, K.J. | | Deposit date: | 2007-07-01 | | Release date: | 2008-05-20 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction.
Nature, 453, 2008
|
|
4F1S
 
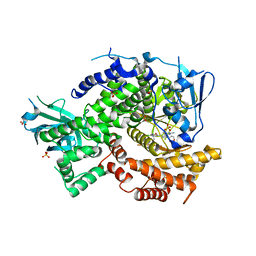 | | Crystal structure of human PI3K-gamma in complex with a pyridyl-triazine-sulfonamide inhibitor | | Descriptor: | N-(5-{[3-(4-amino-6-methyl-1,3,5-triazin-2-yl)-5-(tetrahydro-2H-pyran-4-yl)pyridin-2-yl]amino}-2-chloropyridin-3-yl)methanesulfonamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Whittington, D.A, Tang, J, Yakowec, P. | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationships of dual PI3K/mTOR inhibitors based on a 4-amino-6-methyl-1,3,5-triazine sulfonamide scaffold.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
6DZR
 
 | | Crystal structure of h38C2 K99R mutation | | Descriptor: | CITRATE ANION, SULFATE ION, h38c2 heavy chain, ... | | Authors: | Park, H, Rader, C. | | Deposit date: | 2018-07-05 | | Release date: | 2019-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Site-Selective Antibody Functionalization via Orthogonally Reactive Arginine and Lysine Residues.
Cell Chem Biol, 26, 2019
|
|
3QC4
 
 | | PDK1 in complex with DFG-OUT inhibitor xxx | | Descriptor: | 1-(3,4-difluorobenzyl)-2-oxo-N-{(1R)-2-[(2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)oxy]-1-phenylethyl}-1,2-dihydropyridine-3-carboxamide, 3-phosphoinositide-dependent protein kinase 1 | | Authors: | Arndt, J.W. | | Deposit date: | 2011-01-15 | | Release date: | 2011-04-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a potent and highly selective PDK1 inhibitor via fragment-based drug discovery.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
