6W2A
 
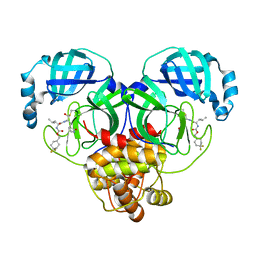 | | 1.65 A resolution structure of SARS-CoV 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, Replicase polyprotein 1a, [4,4-bis(fluoranyl)cyclohexyl]methyl ~{N}-[(2~{S})-1-[[(1~{R},2~{S})-1-[bis(oxidanyl)-oxidanylidene-$l^{5}-sulfanyl]-1-oxidanyl-3-[(3~{S})-2-oxidanylidenepyrrolidin-3-yl]propan-2-yl]amino]-4-methyl-1-oxidanylidene-pentan-2-yl]carbamate | | Authors: | Kashipathy, M.M, Lovell, S, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
6W63
 
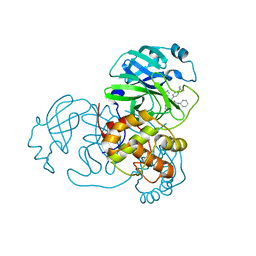 | |
1JKY
 
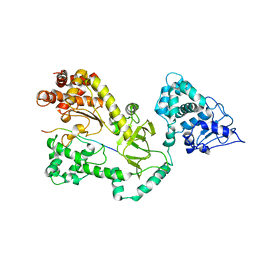 | | Crystal Structure of the Anthrax Lethal Factor (LF): Wild-type LF Complexed with the N-terminal Sequence of MAPKK2 | | Descriptor: | Lethal Factor, mitogen-activated protein kinase kinase 2 | | Authors: | Pannifer, A.D, Wong, T.Y, Schwarzenbacher, R, Renatus, M, Petosa, C, Collier, R.J, Bienkowska, J, Lacy, D.B, Park, S, Leppla, S.H, Hanna, P, Liddington, R.C. | | Deposit date: | 2001-07-13 | | Release date: | 2001-11-07 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the anthrax lethal factor.
Nature, 414, 2001
|
|
6W5K
 
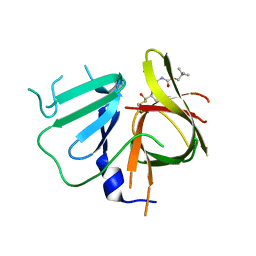 | | 1.95 A resolution structure of Norovirus 3CL protease in complex with inhibitor 5g | | Descriptor: | 3C-LIKE PROTEASE, N~2~-{[2-(3-chlorophenyl)-2-methylpropoxy]carbonyl}-N-{(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}-L-leucinamide | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
6W81
 
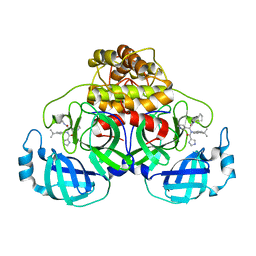 | |
1RU9
 
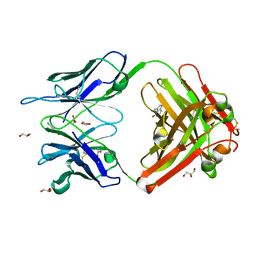 | | Crystal Structure (A) of u.v.-irradiated cationic cyclization antibody 4C6 Fab at pH 4.6 with a data set collected in-house. | | Descriptor: | ACETATE ION, BENZOIC ACID, GLYCEROL, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
1RUK
 
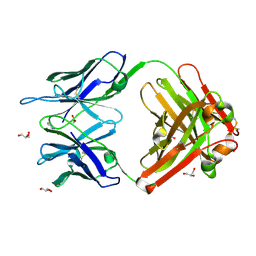 | | Crystal structure (C) of native cationic cyclization antibody 4C6 fab at pH 4.6 with a data set collected at SSRL beamline 9-1 | | Descriptor: | ACETATE ION, BENZOIC ACID, GLYCEROL, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
6WCO
 
 | |
1R4Q
 
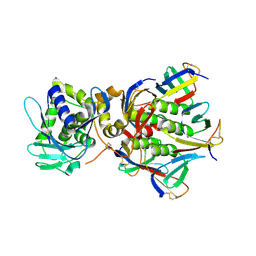 | | Shiga toxin | | Descriptor: | SHT cytotoxin A subunit, Shigella toxin chain B | | Authors: | Fraser, M.E, Fujinaga, M, Cherney, M.M, Melton-Celsa, A.R, Twiddy, E.M, O'Brien, A.D, James, M.N.G. | | Deposit date: | 2003-10-07 | | Release date: | 2004-05-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of Shiga Toxin Type 2 (Stx2) from Escherichia coli O157:H7.
J.Biol.Chem., 279, 2004
|
|
1RUM
 
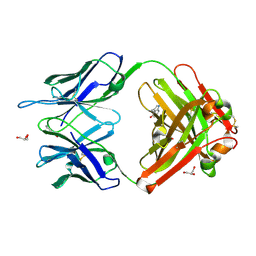 | | Crystal structure (F) of H2O2-soaked cationic cyclization antibody 4C6 fab at pH 8.5 with a data set collected at SSRL beamline 9-1. | | Descriptor: | BENZOIC ACID, GLYCEROL, immunoglobulin igg2a, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
1JFP
 
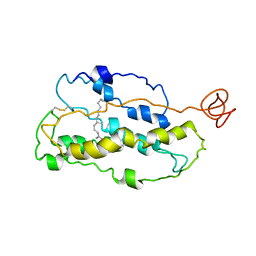 | | Structure of bovine rhodopsin (dark adapted) | | Descriptor: | RETINAL, rhodopsin | | Authors: | Yeagle, P.L, Choi, G, Albert, A.D. | | Deposit date: | 2001-06-21 | | Release date: | 2001-10-05 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Studies on the structure of the G-protein-coupled receptor rhodopsin including the putative G-protein binding site in unactivated and activated forms.
Biochemistry, 40, 2001
|
|
1KEP
 
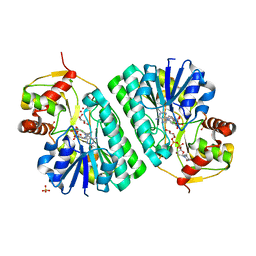 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Streptococcus suis with dTDP-xylose bound | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, THYMIDINE-5'-DIPHOSPHO-BETA-D-XYLOSE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-16 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
1RUA
 
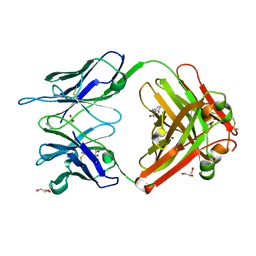 | | Crystal structure (B) of u.v.-irradiated cationic cyclization antibody 4C6 fab at pH 4.6 with a data set collected at SSRL beamline 11-1. | | Descriptor: | ACETATE ION, BENZOIC ACID, GLYCEROL, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
1RUP
 
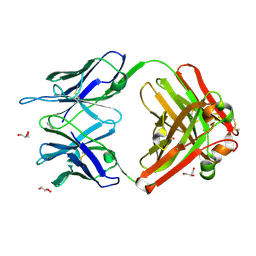 | | Crystal structure (G) of native cationic cyclization antibody 4C6 fab at pH 8.5 with a data set collected at APS beamline 19-ID | | Descriptor: | BENZOIC ACID, GLYCEROL, immunoglobulin igg2a, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
3S7F
 
 | |
1KEW
 
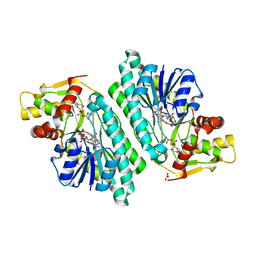 | | The crystal structure of dTDP-D-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium with thymidine diphosphate bound | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Allard, S.T.M, Beis, K, Giraud, M.-F, Hegeman, A.D, Gross, J.W, Whitfield, C, Graninger, M, Messner, P, Allen, A.G, Naismith, J.H. | | Deposit date: | 2001-11-17 | | Release date: | 2002-01-25 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a structural understanding of the dehydratase mechanism.
Structure, 10, 2002
|
|
6WIN
 
 | | Type 6 secretion amidase effector 2 (Tae2) | | Descriptor: | Type 6 secretion amidase effector 2 | | Authors: | Chou, S, Radkov, A.D. | | Deposit date: | 2020-04-10 | | Release date: | 2020-04-22 | | Last modified: | 2021-05-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Ticks Resist Skin Commensals with Immune Factor of Bacterial Origin.
Cell, 183, 2020
|
|
1R8P
 
 | | HPV-16 E2C solution structure | | Descriptor: | Regulatory protein E2 | | Authors: | Nadra, A.D, Eliseo, T, Cicero, D.O. | | Deposit date: | 2003-10-28 | | Release date: | 2004-11-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HPV-16 E2 DNA binding domain, a transcriptional regulator with a dimeric beta-barrel fold
J.Biomol.NMR, 30, 2004
|
|
1UJZ
 
 | | Crystal structure of the E7_C/Im7_C complex; a computationally designed interface between the colicin E7 DNase and the Im7 Immunity protein | | Descriptor: | Designed Colicin E7 DNase, Designed Colicin E7 immunity protein | | Authors: | Kortemme, T, Joachimiak, L.A, Bullock, A.N, Schuler, A.D, Stoddard, B.L, Baker, D. | | Deposit date: | 2003-08-13 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Computational redesign of protein-protein interaction specificity
NAT.STRUCT.MOL.BIOL., 11, 2004
|
|
6W79
 
 | |
1RUR
 
 | | Crystal Structure (I) of native Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected at SSRL beamline 9-1. | | Descriptor: | ZINC ION, immunoglobulin 13G5, heavy chain, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
1RUQ
 
 | | Crystal Structure (H) of u.v.-irradiated Diels-Alder antibody 13G5 Fab at pH 8.0 with a data set collected in house. | | Descriptor: | ZINC ION, immunoglobulin 13G5 heavy chain, immunoglobulin 13G5 light chain | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
6W5J
 
 | | 1.85 A resolution structure of Norovirus 3CL protease in complex with inhibitor 7d | | Descriptor: | 2-(3-chlorophenyl)-2-methylpropyl [(2S)-3-cyclohexyl-1-({(1R,2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]-1-sulfanylpropan-2-yl}amino)-1-oxopropan-2-yl]carbamate, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Kim, Y, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-03-13 | | Release date: | 2020-09-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 63, 2020
|
|
1RUL
 
 | | Crystal Structure (D) of u.v.-irradiated cationic cyclization antibody 4C6 Fab at pH 5.6 with a data set collected at SSRL beamline 11-1. | | Descriptor: | ACETATE ION, BENZOIC ACID, GLYCEROL, ... | | Authors: | Zhu, X, Wentworth Jr, P, Wentworth, A.D, Eschenmoser, A, Lerner, R.A, Wilson, I.A. | | Deposit date: | 2003-12-11 | | Release date: | 2004-03-02 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Probing the antibody-catalyzed water-oxidation pathway at atomic resolution.
Proc.Natl.Acad.Sci.USA, 110, 2004
|
|
3S26
 
 | | Crystal Structure of Murine Siderocalin (Lipocalin 2, 24p3) | | Descriptor: | Neutrophil gelatinase-associated lipocalin, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Correnti, C, Bandaranayake, A.D, Strong, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-28 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors.
Nucleic Acids Res., 39, 2011
|
|
