7B7V
 
 | | Structure of NUDT15 in complex with Acyclovir monophosphate | | Descriptor: | 2-[(2-amino-6-oxo-1,6-dihydro-9H-purin-9-yl)methoxy]ethyl dihydrogen phosphate, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Rehling, D, Stenmark, P. | | Deposit date: | 2020-12-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | NUDT15 polymorphism influences the metabolism and therapeutic effects of acyclovir and ganciclovir.
Nat Commun, 12, 2021
|
|
2R5Y
 
 | | Structure of Scr/Exd complex bound to a consensus Hox-Exd site | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DCP*DTP*DAP*DTP*DGP*DAP*DTP*DTP*DTP*DAP*DTP*DGP*DGP*DGP*DCP*DTP*DG)-3'), DNA (5'-D(*DTP*DCP*DAP*DGP*DCP*DCP*DCP*DAP*DTP*DAP*DAP*DAP*DTP*DCP*DAP*DTP*DAP*DGP*DAP*DG)-3'), Homeobox protein extradenticle, ... | | Authors: | Aggarwal, A.K, Passner, J.M, Jain, R. | | Deposit date: | 2007-09-04 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional specificity of a Hox protein mediated by the recognition of minor groove structure
Cell(Cambridge,Mass.), 131, 2007
|
|
2R5Z
 
 | | Structure of Scr/Exd complex bound to a DNA sequence derived from the fkh gene | | Descriptor: | DNA (5'-D(*DAP*DCP*DTP*DCP*DTP*DAP*DAP*DGP*DAP*DTP*DTP*DAP*DAP*DTP*DCP*DGP*DGP*DCP*DTP*DG)-3'), DNA (5'-D(*DTP*DCP*DAP*DGP*DCP*DCP*DGP*DAP*DTP*DTP*DAP*DAP*DTP*DCP*DTP*DTP*DAP*DGP*DAP*DG)-3'), Homeobox protein extradenticle, ... | | Authors: | Aggarwal, A.K, Passner, J.M, Jain, R. | | Deposit date: | 2007-09-04 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Functional specificity of a Hox protein mediated by the recognition of minor groove structure
Cell(Cambridge,Mass.), 131, 2007
|
|
5Y4Q
 
 | | Crystal structure of Trypanosoma cruzi spermidine synthase in complex with N-(4-methoxyphenyl)quinolin-4-amine | | Descriptor: | 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, N-(4-methoxyphenyl)quinolin-4-amine, Spermidine synthase, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of a Hidden Trypanosoma cruzi Spermidine Synthase Binding Site and Inhibitors through In Silico, In Vitro , and X-ray Crystallography.
Acs Omega, 8, 2023
|
|
5Y4P
 
 | | Crystal structure of Trypanosoma cruzi spermidine synthase in complex with 5-methoxy-2-(5-methyl-4,5-dihydro-1H-imidazol-2-yl)phenol | | Descriptor: | 5'-[(S)-(3-AMINOPROPYL)(METHYL)-LAMBDA~4~-SULFANYL]-5'-DEOXYADENOSINE, 5-methoxy-2-[(5R)-5-methyl-4,5-dihydro-1H-imidazol-2-yl]phenol, Spermidine synthase, ... | | Authors: | Amano, Y, Tateishi, Y. | | Deposit date: | 2017-08-04 | | Release date: | 2018-08-08 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Discovery of a Hidden Trypanosoma cruzi Spermidine Synthase Binding Site and Inhibitors through In Silico, In Vitro , and X-ray Crystallography.
Acs Omega, 8, 2023
|
|
3JVW
 
 | | HIV-1 Protease Mutant G86A with symmetric inhibitor DMP323 | | Descriptor: | Gag-Pol polyprotein, [4-R-(-4-ALPHA,5-ALPHA,6-BETA,7-BETA)]-HEXAHYDRO-5,6-BIS(HYDROXY)-[1,3-BIS([4-HYDROXYMETHYL-PHENYL]METHYL)-4,7-BIS(PHEN YLMETHYL)]-2H-1,3-DIAZEPINONE | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JW2
 
 | | HIV-1 Protease Mutant G86S with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
3JVY
 
 | | HIV-1 Protease Mutant G86A with DARUNAVIR | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, CHLORIDE ION, Gag-Pol polyprotein, ... | | Authors: | Tie, Y, Weber, I.T. | | Deposit date: | 2009-09-17 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Highly conserved glycine 86 and arginine 87 residues contribute differently to the structure and activity of the mature HIV-1 protease
Proteins, 78, 2009
|
|
2LF4
 
 | |
7W3O
 
 | | Crystal structure of human CYB5R3 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADH-cytochrome b5 reductase 3 soluble form | | Authors: | Noda, N.N. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
Nat Commun, 13, 2022
|
|
7W3N
 
 | | Crystal structure of Ufm1 fused to UFBP1 UFIM | | Descriptor: | UFBP1 peptide,Ubiquitin-fold modifier 1 | | Authors: | Noda, N.N. | | Deposit date: | 2021-11-25 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The UFM1 system regulates ER-phagy through the ufmylation of CYB5R3.
Nat Commun, 13, 2022
|
|
5B73
 
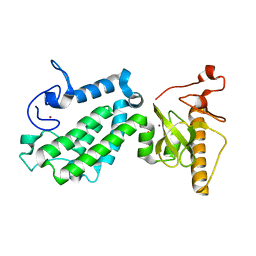 | | Crystal structure of human ZMYND8 PHD-Bromo-PWWP domain | | Descriptor: | Protein kinase C-binding protein 1, ZINC ION | | Authors: | Li, H, Li, Y, Zheng, X. | | Deposit date: | 2016-06-03 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | ZMYND8 Reads the Dual Histone Mark H3K4me1-H3K14ac to Antagonize the Expression of Metastasis-Linked Genes
Mol.Cell, 63, 2016
|
|
5CLB
 
 | |
5CL3
 
 | |
5CL8
 
 | |
5CLE
 
 | |
5CL9
 
 | |
5CL4
 
 | |
5CLA
 
 | |
5CL7
 
 | |
5CLD
 
 | |
5CL5
 
 | |
5CL6
 
 | |
5CLC
 
 | |
5K56
 
 | | Human muscle fructose-1,6-bisphosphatase in active R-state in complex with fructose-1,6-bisphosphate | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, Fructose-1,6-bisphosphatase isozyme 2 | | Authors: | Barciszewski, J, Wisniewski, J, Kolodziejczyk, R, Dzugaj, A, Jaskolski, M, Rakus, D. | | Deposit date: | 2016-05-23 | | Release date: | 2017-06-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural studies of human muscle FBPase
To Be Published
|
|
