2FCS
 
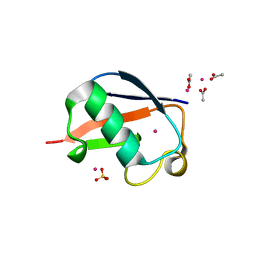 | | X-ray Crystal Structure of a Chemically Synthesized [L-Gln35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, SULFATE ION, ... | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
8DFA
 
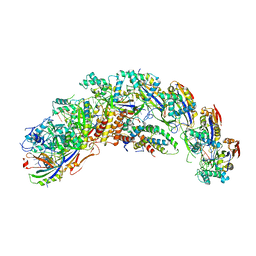 | | type I-C Cascade bound to ssDNA target | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
2FG6
 
 | | N-succinyl-L-ornithine transcarbamylase from B. fragilis complexed with sulfate and N-succinyl-L-norvaline | | Descriptor: | N-(3-CARBOXYPROPANOYL)-L-NORVALINE, SULFATE ION, putative ornithine carbamoyltransferase | | Authors: | Shi, D, Yu, X, Malamy, M.H, Allewell, N.M, Mendel, T. | | Deposit date: | 2005-12-21 | | Release date: | 2006-05-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic mechanism of a novel N-succinyl-L-ornithine transcarbamylase in arginine biosynthesis of Bacteroides fragilis.
J.Biol.Chem., 281, 2006
|
|
8DEX
 
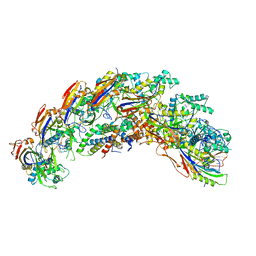 | | type I-C Cascade | | Descriptor: | CRISPR-associated protein, CT1133 family, TM1801 family, ... | | Authors: | O'Brien, R.E, Bravo, J.P.K, Ramos, D, Hibshman, G.N, Wright, J.T, Taylor, D.W. | | Deposit date: | 2022-06-21 | | Release date: | 2023-02-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural snapshots of R-loop formation by a type I-C CRISPR Cascade.
Mol.Cell, 83, 2023
|
|
2FLR
 
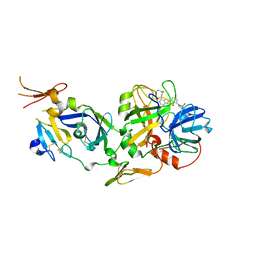 | | Novel 5-Azaindole Factor VIIa Inhibitors | | Descriptor: | Coagulation factor VII, Tissue factor, [2'-HYDROXY-3'-(1H-PYRROLO[3,2-C]PYRIDIN-2-YL)-BIPHENYL-3-YLMETHYL]-UREA | | Authors: | Riggs, J.R, Hu, H, Kolesnikov, A, Tong, Z, Leahy, E.M, Wesson, K.E, Shrader, W.D, Vijaykumar, D, Wahl, T.A, Sprengeler, P.A, Green, M.J, Yu, C, Katz, B.A, Young, W.B. | | Deposit date: | 2006-01-06 | | Release date: | 2007-01-23 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Novel 5-azaindole factor VIIa inhibitors.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2FMG
 
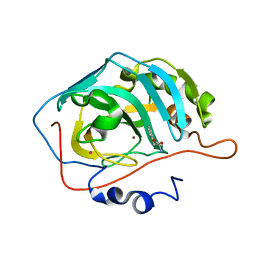 | | Carbonic anhydrase activators. Activation of isoforms I, II, IV, VA, VII and XIV with L- and D- phenylalanine and crystallographic analysis of their adducts with isozyme II: sterospecific recognition within the active site of an enzyme and its consequences for the drug design, structure with L-phenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURY (II) ION, PHENYLALANINE, ... | | Authors: | Temperini, C, Scozzafava, A, Vullo, D, Supuran, C.T. | | Deposit date: | 2006-01-09 | | Release date: | 2006-05-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Carbonic Anhydrase Activators. Activation of Isoforms I, II, IV, VA, VII, and XIV with l- and d-Phenylalanine and Crystallographic Analysis of Their Adducts with Isozyme II: Stereospecific Recognition within the Active Site of an Enzyme and Its Consequences for the Drug Design.
J.Med.Chem., 49, 2006
|
|
1VT9
 
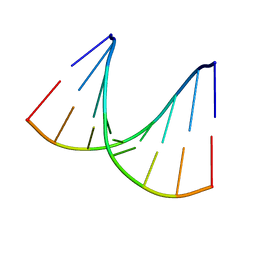 | |
1VL3
 
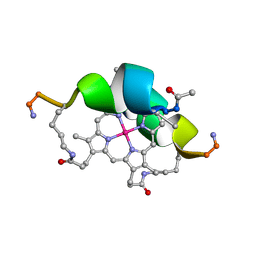 | | DESIGN OF NEW MIMOCHROMES WITH UNIQUE TOPOLOGY | | Descriptor: | CO(III)-(DEUTEROPORPHYRIN IX), GLU-SER-GLN-LEU-HIS-SER-ASN-LYS-ARG | | Authors: | Lombardi, A, Nastri, F, Marasco, D, Maglio, O, De Sanctis, G, Sinibaldi, F, Santucci, R, Coletta, M, Pavone, V. | | Deposit date: | 2004-07-05 | | Release date: | 2004-07-20 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Design of a New Mimochrome with Unique Topology.
Chemistry, 9, 2003
|
|
1VZI
 
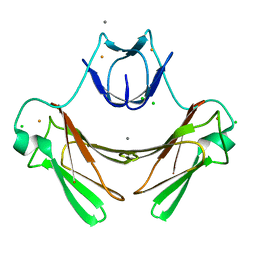 | | Structure of superoxide reductase bound to ferrocyanide and active site expansion upon X-ray induced photoreduction | | Descriptor: | CALCIUM ION, CHLORIDE ION, DESULFOFERRODOXIN, ... | | Authors: | Adam, V, Royant, A, Niviere, V, Molina-Heredia, F.P, Bourgeois, D. | | Deposit date: | 2004-05-19 | | Release date: | 2004-08-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Structure of superoxide reductase bound to ferrocyanide and active site expansion upon X-ray-induced photo-reduction.
Structure, 12, 2004
|
|
2EZ9
 
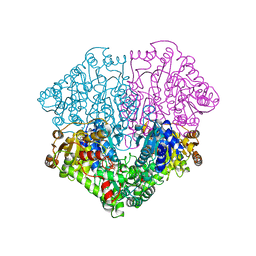 | | Pyruvate oxidase variant F479W in complex with reaction intermediate analogue 2-phosphonolactyl-thiamin diphosphate | | Descriptor: | 3-[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]-2-{(1S)-1-HYDROXY-1-[(R)-HYDROXY(METHOXY)PHOSPHORYL]ETHYL}-5-(2-{[(S)-HYDROXY(PHOSPHONOOXY)PHOSPHORYL]OXY}ETHYL)-4-METHYL-1,3-THIAZOL-3-IUM, FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, ... | | Authors: | Wille, G, Meyer, D, Steinmetz, A, Hinze, E, Golbik, R, Tittmann, K. | | Deposit date: | 2005-11-10 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The catalytic cycle of a thiamin diphosphate enzyme examined by cryocrystallography.
Nat.Chem.Biol., 2, 2006
|
|
1W5W
 
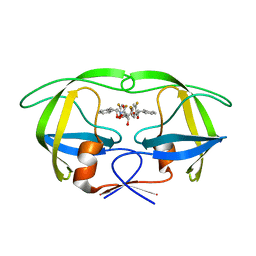 | | HIV-1 protease in complex with fluoro substituted diol-based C2- symmetric inhibitor | | Descriptor: | (2R,3R,4R,5R)-2,5-BIS[(2,4-DIFLUOROBENZYL)OXY]-3,4-DIHYDROXY-N,N'-BIS[(1R,2S)-2-HYDROXY-2,3-DIHYDRO-1H-INDEN-1-YL]HEXAN EDIAMIDE, POL POLYPROTEIN | | Authors: | Lindberg, J, Pyring, D, Loewgren, S, Rosenquist, A, Zuccarello, G, Kvarnstroem, I, Zhang, H, Vrang, L, Claesson, B, Hallberg, A, Samuelsson, B, Unge, T. | | Deposit date: | 2004-08-10 | | Release date: | 2004-12-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Symmetric Fluoro-Substituted Diol-Based HIV Protease Inhibitors. Ortho-Fluorinated and Meta-Fluorinated P1/P1'-Benzyloxy Side Groups Significantly Improve the Antiviral Activity and Preserve Binding Efficacy
Eur.J.Biochem., 271, 2004
|
|
1VEL
 
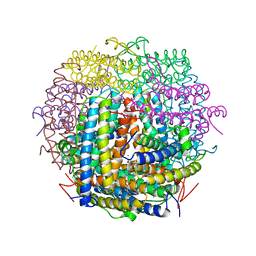 | | Mycobacterium smegmatis Dps tetragonal form | | Descriptor: | CADMIUM ION, SODIUM ION, SULFATE ION, ... | | Authors: | Roy, S, Gupta, S, Das, S, Sekar, K, Chatterji, D, Vijayan, M. | | Deposit date: | 2004-04-01 | | Release date: | 2004-06-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules
J.Mol.Biol., 339, 2004
|
|
1W9H
 
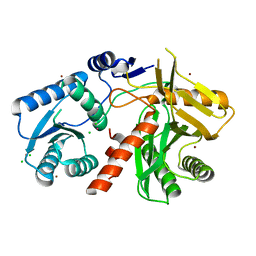 | | The Structure of a Piwi protein from Archaeoglobus fulgidus. | | Descriptor: | CADMIUM ION, CHLORIDE ION, HYPOTHETICAL PROTEIN AF1318, ... | | Authors: | Parker, J.S, Roe, S.M, Barford, D. | | Deposit date: | 2004-10-13 | | Release date: | 2005-01-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a Piwi Protein Suggests Mechanisms for Sirna Recognition and Slicer Activity
Embo J., 23, 2004
|
|
2F73
 
 | | Crystal structure of human fatty acid binding protein 1 (FABP1) | | Descriptor: | Fatty acid-binding protein, liver | | Authors: | Kursula, P, Thorsell, A.G, Arrowsmith, C, Berglund, H, Edwards, A, Ehn, M, Flodin, S, Graslund, S, Hammarstrom, M, Holmberg Schiavone, L, Kotenyova, T, Nilsson-Ehle, P, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, van den Berg, S, Weigelt, J, Hallberg, B.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-11-30 | | Release date: | 2005-12-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human FABP1
To be Published
|
|
2F7V
 
 | | Structure of acetylcitrulline deacetylase complexed with one Co | | Descriptor: | COBALT (II) ION, aectylcitrulline deacetylase | | Authors: | Shi, D, Yu, X, Roth, L, Allewell, N.M, Tuchman, M. | | Deposit date: | 2005-12-01 | | Release date: | 2006-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a novel N-acetyl-L-citrulline deacetylase from Xanthomonas campestris
Biophys.Chem., 126, 2007
|
|
1VJM
 
 | | Deformation of helix C in the low-temperature L-intermediate of bacteriorhodopsin | | Descriptor: | Bacteriorhodopsin, RETINAL | | Authors: | Edman, K, Royant, A, Larsson, G, Jacobson, F, Taylor, T, van der Spoel, D, Landau, E.M, Pebay-Peyroula, E, Neutze, R. | | Deposit date: | 2004-03-12 | | Release date: | 2004-04-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Deformation of helix C in the low temperature L-intermediate of bacteriorhodopsin.
J.Biol.Chem., 279, 2004
|
|
2FCN
 
 | | X-ray Crystal Structure of a Chemically Synthesized [D-Val35]Ubiquitin with a Cubic Space Group | | Descriptor: | ACETATE ION, CADMIUM ION, Ubiquitin | | Authors: | Bang, D, Gribenko, A.V, Tereshko, V, Kossiakoff, A.A, Kent, S.B, Makhatadze, G.I. | | Deposit date: | 2005-12-12 | | Release date: | 2006-01-31 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dissecting the energetics of protein alpha-helix C-cap termination through chemical protein synthesis.
Nat.Chem.Biol., 2, 2006
|
|
2FGG
 
 | | Crystal Structure of Rv2632c | | Descriptor: | Hypothetical protein Rv2632c/MT2708 | | Authors: | Yu, M, Bursey, E.H, Radhakannan, T, Segelke, B.W, Lekin, T, Toppani, D, Kim, C.Y, Kaviratne, T, Woodruff, T, Terwilliger, T.C, Hung, L.W, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-12-21 | | Release date: | 2006-02-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Rv2632c
To be Published
|
|
2FHB
 
 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FGZ
 
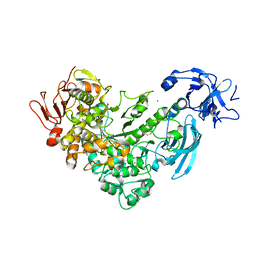 | | Crystal Structure Analysis of apo pullulanase from Klebsiella pneumoniae | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
2FHF
 
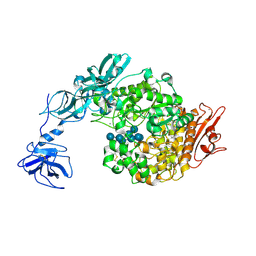 | | Crystal Structure Analysis of Klebsiella pneumoniae pullulanase complexed with maltotetraose | | Descriptor: | Alpha-dextrin endo-1,6-alpha-glucosidase, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Mikami, B, Iwamoto, H, Katsuya, Y, Yoon, H.-J, Demirkan-Sarikaya, E, Malle, D. | | Deposit date: | 2005-12-23 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of pullulanase: evidence for parallel binding of oligosaccharides in the active site
J.Mol.Biol., 359, 2006
|
|
1VSR
 
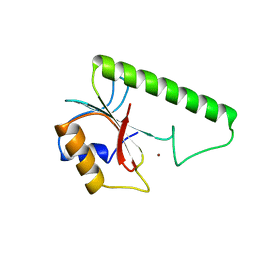 | | VERY SHORT PATCH REPAIR (VSR) ENDONUCLEASE FROM ESCHERICHIA COLI | | Descriptor: | PROTEIN (VSR ENDONUCLEASE), ZINC ION | | Authors: | Tsutakawa, S.E, Muto, T, Jingami, H, Kunishima, N, Ariyoshi, M, Kohda, D, Nakagawa, M, Morikawa, K. | | Deposit date: | 1999-02-13 | | Release date: | 1999-10-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallographic and functional studies of very short patch repair endonuclease.
Mol.Cell, 3, 1999
|
|
1VT8
 
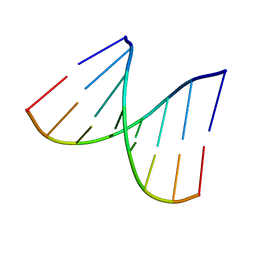 | | Crystal structure of D(GGGCGCCC)-hexagonal form | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1996-12-02 | | Release date: | 2011-07-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
2FM0
 
 | | Crystal structure of PDE4D in complex with L-869298 | | Descriptor: | (S)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHY L)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-06 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
2FM5
 
 | | Crystal structure of PDE4D2 in complex with inhibitor L-869299 | | Descriptor: | (R)-3-(2-(3-CYCLOPROPOXY-4-(DIFLUOROMETHOXY)PHENYL)-2-(5-(1,1,1,3,3,3-HEXAFLUORO-2-HYDROXYPROPAN-2-YL)THIAZOL-2-YL)ETHYL)PYRIDINE 1-OXIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Huai, Q, Sun, Y, Wang, H, Macdonald, D, Aspiotis, R, Robinson, H, Huang, Z, Ke, H. | | Deposit date: | 2006-01-07 | | Release date: | 2006-03-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Enantiomer Discrimination Illustrated by the High Resolution Crystal Structures of Type 4 Phosphodiesterase
J.Med.Chem., 49, 2006
|
|
