7SXR
 
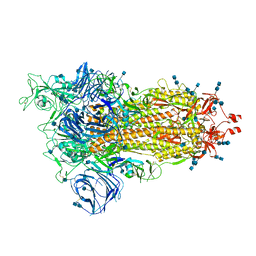 | | Cryo-EM structure of the SARS-CoV-2 D614G mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXV
 
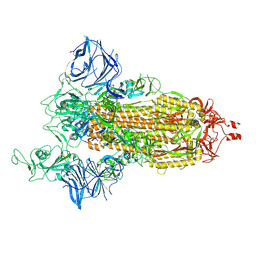 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K,K417N mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (2.79 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
7SXU
 
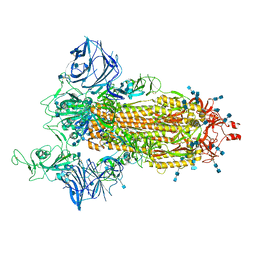 | | Cryo-EM structure of the SARS-CoV-2 D614G,N501Y,E484K mutant spike protein ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Mannar, D, Saville, J.W, Srivastava, S.S, Berezuk, A.M, Zhou, S, Tuttle, K.S, Kim, A, Li, W, Dimitrov, D.S, Subramaniam, S. | | Deposit date: | 2021-11-24 | | Release date: | 2021-12-29 | | Last modified: | 2022-01-05 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural analysis of receptor binding domain mutations in SARS-CoV-2 variants of concern that modulate ACE2 and antibody binding.
Cell Rep, 37, 2021
|
|
8UM3
 
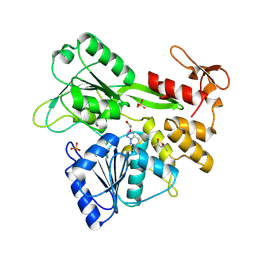 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
8UBH
 
 | |
8U45
 
 | | Crystal Structure Analysis of Aspergillus fumigatus alkaline protease | | Descriptor: | Alkaline protease 1, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Diec, D.D.L, Guo, W, Russi, S. | | Deposit date: | 2023-09-08 | | Release date: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Aspergillus allergen oryzin with a chemical probe at atomic precision.
Sci Rep, 13, 2023
|
|
3A99
 
 | | Structure of PIM-1 kinase crystallized in the presence of P27KIP1 Carboxy-terminal peptide | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Proto-oncogene serine/threonine-protein kinase pim-1 | | Authors: | Morishita, D, Takami, M, Yoshikawa, S, Katayama, R, Sato, S, Kukimoto-Niino, M, Umehara, T, Shirouzu, M, Sekimizu, K, Yokoyama, S, Fujita, N. | | Deposit date: | 2009-10-22 | | Release date: | 2010-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Cell-permeable carboxyl-terminal p27(Kip1) peptide exhibits anti-tumor activity by inhibiting Pim-1 kinase
J.Biol.Chem., 286, 2011
|
|
3AH5
 
 | | Crystal Structure of flavin dependent thymidylate synthase ThyX from helicobacter pylori complexed with FAD and dUMP | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Zhang, X, Zhang, J, Hu, Y, Zou, Q, Wang, D. | | Deposit date: | 2010-04-14 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and functional analysis of a flavin dependent thymidylate synthase from helicobacter pylori
To be Published
|
|
3AB1
 
 | | Crystal Structure of Ferredoxin NADP+ Oxidoreductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Ferredoxin--NADP reductase | | Authors: | Muraki, N, Seo, D, Kurisu, G. | | Deposit date: | 2009-11-30 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Asymmetric dimeric structure of ferredoxin-NAD(P)+ oxidoreductase from the green sulfur bacterium Chlorobaculum tepidum: implications for binding ferredoxin and NADP+
J.Mol.Biol., 401, 2010
|
|
2E4Z
 
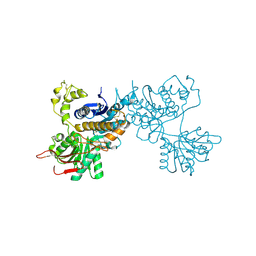 | | Crystal structure of the ligand-binding region of the group III metabotropic glutamate receptor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Metabotropic glutamate receptor 7 | | Authors: | Muto, T, Tsuchiya, D, Morikawa, K, Jingami, H. | | Deposit date: | 2006-12-17 | | Release date: | 2007-02-27 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structures of the extracellular regions of the group II/III metabotropic glutamate receptors
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
7ZPJ
 
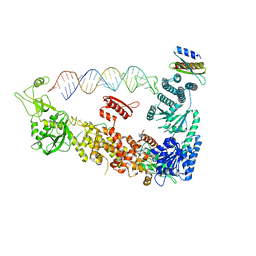 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 isoform 1 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
3AEM
 
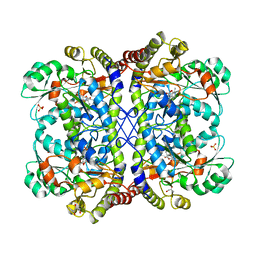 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing Michaelis complex and methionine imine-pyridoxamine-5'-phosphate | | Descriptor: | (2E)-2-[({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)imino]-4-(methylsulfanyl)butanoic acid, GLYCEROL, METHIONINE, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be published
|
|
307D
 
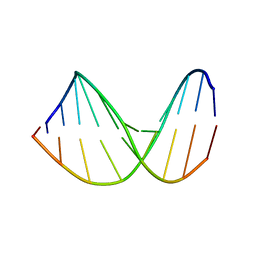 | | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis | | Descriptor: | DNA (5'-D(*CP*AP*AP*AP*GP*AP*AP*AP*AP*G)-3'), DNA (5'-D(*CP*TP*TP*TP*TP*CP*TP*TP*TP*G)-3') | | Authors: | Han, G.W, Kopka, M.L, Cascio, D, Grzeskowiak, K, Dickerson, R.E. | | Deposit date: | 1997-01-07 | | Release date: | 1997-01-27 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of a DNA analog of the primer for HIV-1 RT second strand synthesis.
J.Mol.Biol., 269, 1997
|
|
7TEZ
 
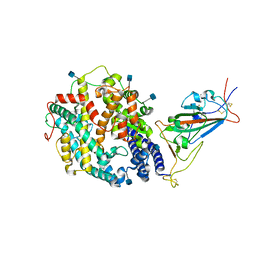 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) spike protein in complex with human ACE2 (focused refinement of RBD and ACE2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
3A4N
 
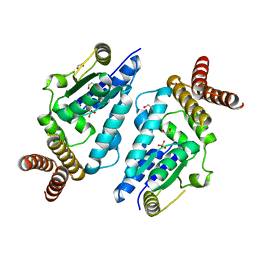 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) kinase | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, L-seryl-tRNA(Sec) kinase, ... | | Authors: | Araiso, Y, Ishitani, R, Soll, D, Nureki, O. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a tRNA-dependent kinase essential for selenocysteine decoding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2JTO
 
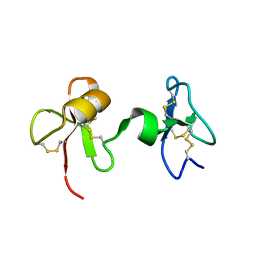 | | Solution Structure of Tick Carboxypeptidase Inhibitor | | Descriptor: | Carboxypeptidase inhibitor | | Authors: | Pantoja-Uceda, D, Blanco, F.J. | | Deposit date: | 2007-08-03 | | Release date: | 2008-07-15 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | The NMR structure and dynamics of the two-domain tick carboxypeptidase inhibitor reveal flexibility in its free form and stiffness upon binding to human carboxypeptidase B.
Biochemistry, 47, 2008
|
|
3AEN
 
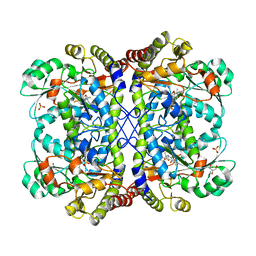 | | Reaction intermediate structure of Entamoeba histolytica methionine gamma-lyase 1 containing Michaelis complex and alpha-amino-alpha, beta-butenoic acid-pyridoxal-5'-phosphate | | Descriptor: | (2E)-2-{[(1E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}but-2-enoic acid, GLYCEROL, METHIONINE, ... | | Authors: | Karaki, T, Sato, D, Shimizu, A, Nozaki, T, Harada, S. | | Deposit date: | 2010-02-10 | | Release date: | 2011-02-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of Entamoeba histolytica methionine gamma-lyase 1
To be Published
|
|
3AMA
 
 | | Protein kinase A sixfold mutant model of Aurora B with inhibitor JNJ-7706621 | | Descriptor: | 4-({5-amino-1-[(2,6-difluorophenyl)carbonyl]-1H-1,2,4-triazol-3-yl}amino)benzenesulfonamide, cAMP-dependent protein kinase catalytic subunit alpha, cAMP-dependent protein kinase inhibitor alpha | | Authors: | Pflug, A, de Oliveira, T.M, Bossemeyer, D, Engh, R.A. | | Deposit date: | 2010-08-18 | | Release date: | 2011-08-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Mutants of protein kinase A that mimic the ATP-binding site of Aurora kinase
Biochem.J., 440, 2011
|
|
3AK4
 
 | | Crystal structure of NADH-dependent quinuclidinone reductase from agrobacterium tumefaciens | | Descriptor: | NADH-dependent quinuclidinone reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Miyakawa, T, Kataoka, M, Takeshita, D, Nomoto, F, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of NADH-dependent quinuclidinone reductase from Agrobacterium tumefaciens
To be Published
|
|
7TF2
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) Q484I spike protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
8V1P
 
 | | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092 | | Descriptor: | Glucose-induced degradation protein 4 homolog, N,N~2~-bis[(4-methoxyphenyl)methyl]glycinamide | | Authors: | Dong, C, Dong, A, Calabrese, M, Wang, F, Owen, D, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-21 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | CRYSTAL STRUCTURE OF GID4 IN COMPLEX WITH UBF9092
To be published
|
|
3AVY
 
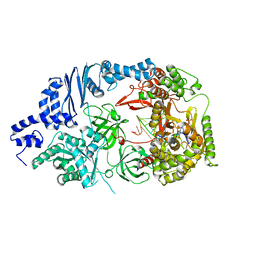 | | Structure of viral RNA polymerase complex 6 | | Descriptor: | 3'-DEOXY-CYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Elongation factor Ts, ... | | Authors: | Takeshita, D, Tomita, K. | | Deposit date: | 2011-03-08 | | Release date: | 2012-01-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.616 Å) | | Cite: | Molecular basis for RNA polymerization by Q beta replicase
Nat.Struct.Mol.Biol., 19, 2012
|
|
7TEX
 
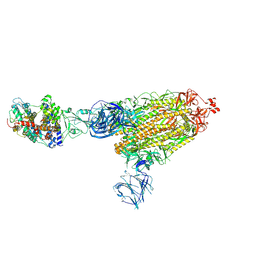 | | Cryo-EM structure of SARS-CoV-2 Delta (B.1.617.2) spike protein in complex with human ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Processed angiotensin-converting enzyme 2, ... | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7TF1
 
 | | Cryo-EM structure of SARS-CoV-2 Kappa (B.1.617.1) Q484I spike protein (focused refinement of RBD) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Zhu, X, Saville, J.W, Mannar, D, Srivastava, S.S, Berezuk, A.M, Demers, J.P, Zhou, S, Tuttle, K.S, Subramaniam, S. | | Deposit date: | 2022-01-06 | | Release date: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Structural and biochemical rationale for enhanced spike protein fitness in delta and kappa SARS-CoV-2 variants.
Nat Commun, 13, 2022
|
|
7ZH2
 
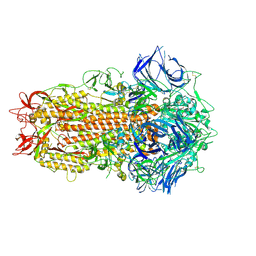 | | SARS CoV Spike protein, Closed C1 conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Buzas, D, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2022-04-05 | | Release date: | 2023-02-15 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | The free fatty acid-binding pocket is a conserved hallmark in pathogenic beta-coronavirus spike proteins from SARS-CoV to Omicron.
Sci Adv, 8, 2022
|
|
