5TKW
 
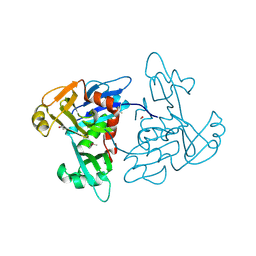 | | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae. | | Descriptor: | FORMIC ACID, Type II secretion system protein L | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-09 | | Release date: | 2016-10-19 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | 1.35 Angstrom Resolution Crystal Structure of a Pullulanase-specific Type II Secretion System Integral Cytoplasmic Membrane Protein GspL (N-terminal fragment; residues 1-237) from Klebsiella pneumoniae.
To Be Published
|
|
5TXG
 
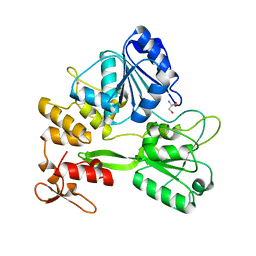 | | Crystal structure of the Zika virus NS3 helicase. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, NS3 helicase, POTASSIUM ION | | Authors: | Nocadello, S, Light, S.H, Minasov, G, Shuvalova, L, Cardona-Correa, A.A, Ojeda, I, Vargas, J, Johnson, M.E, Lee, H, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the Zika virus NS3 helicase.
To be published
|
|
5TVL
 
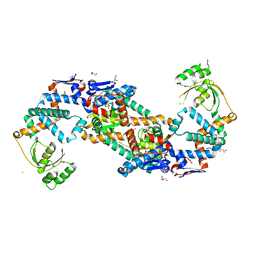 | | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A | | Descriptor: | CHLORIDE ION, Foldase protein PrsA, GLYCEROL, ... | | Authors: | Borek, D, Yim, V, Kudritska, M, Wawrzak, Z, Stogios, P.J, Otwinowski, Z, Savchenko, A, Anderson, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-09 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of foldase protein PrsA from Streptococcus pneumoniae str. Canada MDR_19A
To Be Published
|
|
5TXU
 
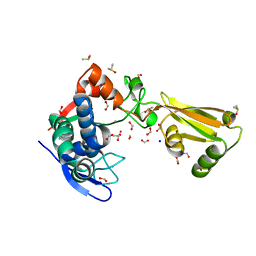 | | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, FORMIC ACID, ... | | Authors: | Nocadello, S, Minasov, G, Kiryukhina, O, Shuvalova, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-17 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Stage II Sporulation Protein D (SpoIID) from Clostridium difficile in Apo Conformation
To Be Published
|
|
5UBU
 
 | | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica. | | Descriptor: | Putative acetamidase/formamidase, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 2.75 Angstrom Resolution Crystal Structure of Acetamidase from Yersinia enterocolitica.
To Be Published
|
|
5TV2
 
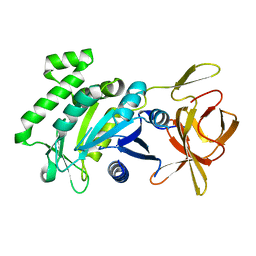 | | Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6 | | Descriptor: | Elongation factor G | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-07 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a fragment (1-405) of an elongation factor G from Vibrio vulnificus CMCP6
To Be Published
|
|
5TTA
 
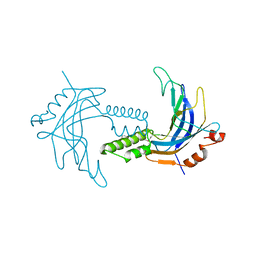 | | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein | | Descriptor: | Putative exported protein | | Authors: | Brunzelle, J.S, Minasov, G, Shuvalova, L, Cordona-Correa, A, Dubrovska, I, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-02 | | Release date: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A 1.85A X-Ray Structure from Peptoclostridium difficile 630 of a Hypothetical Protein
To Be Published
|
|
5U4H
 
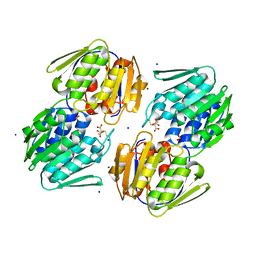 | | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid. | | Descriptor: | (2R)-2-(phosphonooxy)propanoic acid, FORMIC ACID, SODIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-04 | | Release date: | 2016-12-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | 1.05 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Acinetobacter baumannii in Covalently Bound Complex with (2R)-2-(phosphonooxy)propanoic Acid.
To Be Published
|
|
5TW9
 
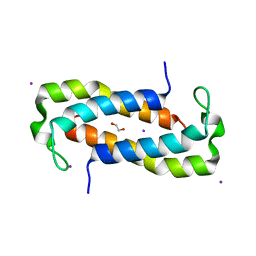 | | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis. | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, Iron uptake system component EfeO | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-11 | | Release date: | 2016-11-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | 1.50 Angstrom Crystal Structure of C-terminal Fragment (residues 322-384) of Iron Uptake System Component EfeO from Yersinia pestis.
To Be Published
|
|
5TV7
 
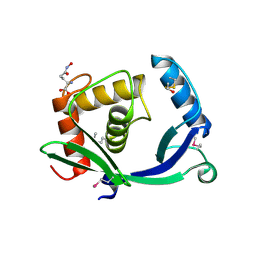 | | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate. | | Descriptor: | GLUTAMINE HYDROXAMATE, Putative peptidoglycan-binding/hydrolysing protein | | Authors: | Minasov, G, Wawrzak, Z, Shuvalova, L, Winsor, J, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-08 | | Release date: | 2016-12-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | 2.05 Angstrom Resolution Crystal Structure of Peptidoglycan-Binding Protein from Clostridioides difficile in Complex with Glutamine Hydroxamate.
To Be Published
|
|
5U47
 
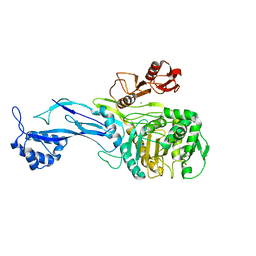 | | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus | | Descriptor: | ACETATE ION, CHLORIDE ION, Penicillin binding protein 2X | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-03 | | Release date: | 2016-12-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Penicillin Binding Protein 2X from Streptococcus thermophilus.
To Be Published
|
|
5UEJ
 
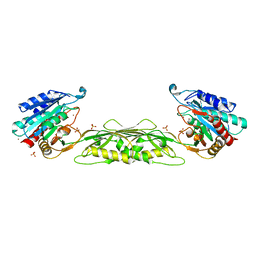 | |
5UE1
 
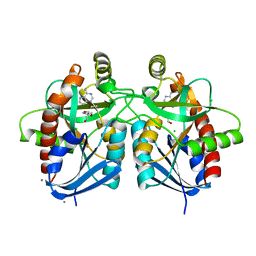 | | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase in complex with adenine from Vibrio fischeri ES114 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase, ... | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-29 | | Release date: | 2017-01-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.14 Å) | | Cite: | Crystal structure of 5'-methylthioadenosine/S-adenosylhomocysteine nucleosidase from Vibrio fischeri ES114
To Be Published
|
|
5UH0
 
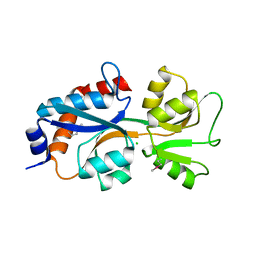 | | 1.95 Angstrom Resolution Crystal Structure of Fragment (35-274) of Membrane-bound Lytic Murein Transglycosylase F from Yersinia pestis. | | Descriptor: | CHLORIDE ION, Membrane-bound lytic murein transglycosylase F | | Authors: | Minasov, G, Shuvalova, L, Flores, K, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-10 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | 1.95 Angstrom Resolution Crystal Structure of Fragment (35-274) of Membrane-bound Lytic Murein Transglycosylase F from Yersinia pestis.
To Be Published
|
|
5UJS
 
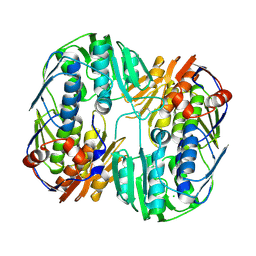 | | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni. | | Descriptor: | CHLORIDE ION, UDP-N-acetylglucosamine 1-carboxyvinyltransferase | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-18 | | Release date: | 2017-02-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | 2.45 Angstrom Resolution Crystal Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Campylobacter jejuni.
To Be Published
|
|
5V10
 
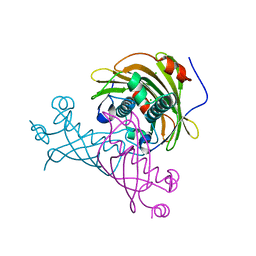 | | Crystal structure of the putative tol-pal system-associated acyl-CoA thioesterase from Pseudomonas aeruginosa PAO1 | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Borek, D, Wawrzak, Z, Grimshaw, S, Sandoval, J, Evdokimova, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative tol-pal system-associated acyl-CoA thioesterase from Pseudomonas aeruginosa PAO1
To Be Published
|
|
5UPU
 
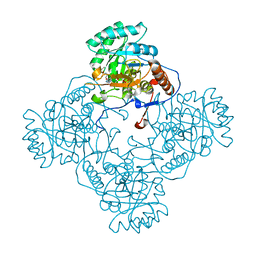 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Kim, Y, Makowska-Grzyska, M, Maltseva, N, Mulligan, R, Gu, M, Sacchettini, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.905 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
5UPY
 
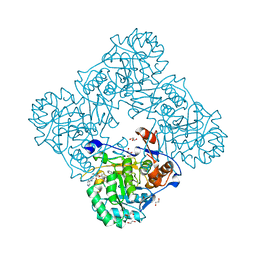 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21 | | Descriptor: | (2S)-2-(naphthalen-1-yloxy)-N-[2-(pyridin-4-yl)-1,3-benzoxazol-5-yl]propanamide, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Osipiuk, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-04 | | Release date: | 2017-04-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Listeria Monocytogenes in the complex with IMP and Q21
To Be Published
|
|
5TPI
 
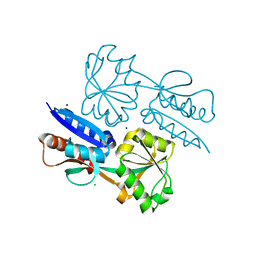 | | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae. | | Descriptor: | CHLORIDE ION, Putative transcriptional regulator (LysR family), SODIUM ION | | Authors: | Minasov, G, Wawrzak, Z, Sandoval, J, Evdokimova, E, Grimshaw, S, Kwon, K, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | 1.47 Angstrom Crystal Structure of the C-terminal Substrate Binding Domain of LysR Family Transcriptional Regulator from Klebsiella pneumoniae.
To Be Published
|
|
5TU0
 
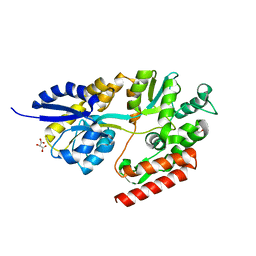 | | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose | | Descriptor: | Lmo2125 protein, TARTRONATE, TRIETHYLENE GLYCOL, ... | | Authors: | Minasov, G, Shuvalova, L, Cardona-Correa, A, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-11-04 | | Release date: | 2016-11-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Maltose-Binding Periplasmic Protein MalE from Listeria monocytogenes in Complex with Maltose.
To Be Published
|
|
5UQH
 
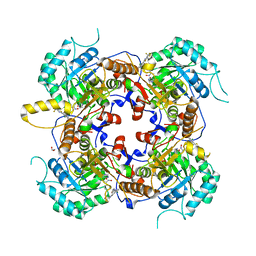 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Campylobacter jejuni in the complex with inhibitor p182 | | Descriptor: | 1,2-ETHANEDIOL, INOSINIC ACID, ISOPROPYL ALCOHOL, ... | | Authors: | Kim, Y, Maltseva, N, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-08 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.201 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Mycobacterium tuberculosis in the presence of TBK6
To Be Published
|
|
5UUV
 
 | | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from Bacillus anthracis in the complex with a product IMP and the inhibitor P182 | | Descriptor: | GLYCEROL, INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Kim, Y, Maltseva, N, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D, Hedstrom, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-17 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal Structure of the Catalytic Domain of the Inosine Monophosphate Dehydrogenase from
Bacillus anthracis in the complex with a product IMP and the inhibitor P182
To Be Published
|
|
5UXE
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from Clostridium perfringens Complexed with IMP and P178 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, FORMIC ACID, INOSINIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Mulligan, R, Makowska-Grzyska, M, Gu, M, Gollapalli, D.R, Hedstrom, L, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase from
Clostridium perfringens
Complexed with IMP and P178
To Be Published
|
|
5UTU
 
 | | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, ADENOSINE, Adenosylhomocysteinase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Stam, J, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | 2.65 Angstrom Resolution Crystal Structure of S-adenosylhomocysteinase from Cryptosporidium parvum in Complex with SAH and NAD
To Be Published
|
|
5UJW
 
 | | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4 | | Descriptor: | 1,2-ETHANEDIOL, 1,3-DIHYDROXYACETONEPHOSPHATE, CITRIC ACID, ... | | Authors: | Chang, C, Maltseva, N, Kim, Y, Shatsman, S, Joachimiak, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-01-19 | | Release date: | 2017-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of triosephosphate isomerase from Francisella tularensis subsp. tularensis SCHU S4
To Be Published
|
|
