3TBS
 
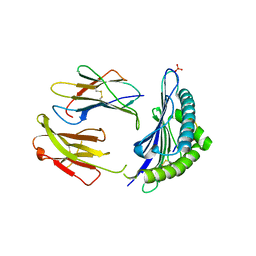 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX THE WITH LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (V3P,Y4A) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, GLYCOPROTEIN G1, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
3RLE
 
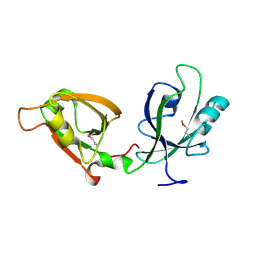 | | Crystal Structure of GRASP55 GRASP domain (residues 7-208) | | Descriptor: | Golgi reassembly-stacking protein 2 | | Authors: | Truschel, S.T, Sengupta, D, Foote, A, Heroux, A, Macbeth, M.R, Linstedt, A.D. | | Deposit date: | 2011-04-19 | | Release date: | 2011-05-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structure of the Membrane-tethering GRASP Domain Reveals a Unique PDZ Ligand Interaction That Mediates Golgi Biogenesis.
J.Biol.Chem., 286, 2011
|
|
2A02
 
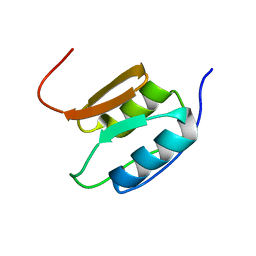 | | Solution NMR Structure of the Periplasmic Signaling Domain of the Outer Membrane Iron Transporter PupA from Pseudomonas putida. | | Descriptor: | Ferric-pseudobactin 358 receptor | | Authors: | Ferguson, A.D, Amezcua, C.A, Chelliah, Y, Rosen, M.K, Deisenhofer, J. | | Deposit date: | 2005-06-15 | | Release date: | 2006-09-26 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Signal transduction pathway of TonB-dependent transporters.
Proc.Natl.Acad.Sci.Usa, 2, 2006
|
|
3TBV
 
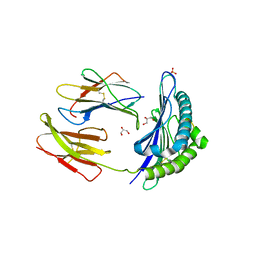 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G,V3P,Y4A) | | Descriptor: | Beta-2-microglobulin, GLYCEROL, Glycoprotein G1, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
1ZZF
 
 | |
3S7J
 
 | |
1XYX
 
 | | mouse prion protein fragment 121-231 | | Descriptor: | Major prion protein | | Authors: | Gossert, A.D, Bonjour, S, Lysek, D.A, Fiorito, F, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of elk and of mouse/elk hybrids
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
2A4H
 
 | | Solution structure of Sep15 from Drosophila melanogaster | | Descriptor: | Selenoprotein Sep15 | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-28 | | Release date: | 2005-12-13 | | Last modified: | 2022-03-09 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
2AAV
 
 | | Solution NMR structure of Filamin A domain 17 | | Descriptor: | Filamin A | | Authors: | Nakamura, F, Pudas, R, Heikkinen, O, Permi, P, Kilpelainen, I, Munday, A.D, Hartwig, J.H, Stossel, T.P, Ylanne, J. | | Deposit date: | 2005-07-14 | | Release date: | 2006-05-02 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The structure of the GPIb-filamin A complex
Blood, 107, 2006
|
|
3TBW
 
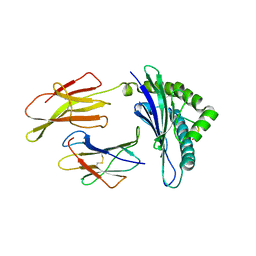 | | CRYSTAL STRUCTURE OF THE MURINE CLASS I MAJOR HISTOCOMPATIBILITY COMPLEX H-2DB IN COMPLEX WITH THE LCMV-DERIVED GP33 ALTERED PEPTIDE ligand (A2G, V3P, Y4S) | | Descriptor: | Beta-2-microglobulin, GLYCOPROTEIN GPC, H-2 class I histocompatibility antigen, ... | | Authors: | Duru, A.D, Allerbring, E.B, Uchtenhagen, H, Mazumdar, P.A, Badia-Martinez, D, Madhurantakam, C, Sandalova, T, Nygren, P, Achour, A. | | Deposit date: | 2011-08-08 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conversion of a T cell viral antagonist into an agonist through higher stabilization and conserved molecular mimicry: Implications for TCR recognition
To be Published
|
|
2A2P
 
 | | Solution structure of SelM from Mus musculus | | Descriptor: | Selenoprotein M | | Authors: | Ferguson, A.D, Labunskyy, V.M, Fomenko, D.E, Chelliah, Y, Amezcua, C.A, Rizo, J, Gladyshev, V.N, Deisenhofer, J. | | Deposit date: | 2005-06-22 | | Release date: | 2005-12-06 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR Structures of the Selenoproteins Sep15 and SelM Reveal Redox Activity of a New Thioredoxin-like Family.
J.Biol.Chem., 281, 2006
|
|
1Y16
 
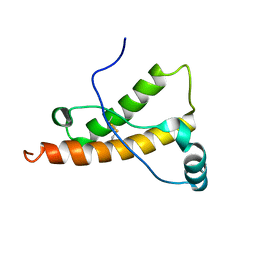 | | mouse prion protein with mutations S170N and N174T | | Descriptor: | Major prion protein | | Authors: | Gossert, A.D, Bonjour, S, Lysek, D.A, Fiorito, F, Wuthrich, K. | | Deposit date: | 2004-11-17 | | Release date: | 2005-01-25 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of elk and of mouse/elk hybrids
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
1Z3R
 
 | | Solution structure of the Omsk Hemhorraghic Fever Envelope Protein Domain III | | Descriptor: | polyprotein | | Authors: | Volk, D.E, Chavez, L, Beasley, D.W, Barrett, A.D, Gorenstein, D.G. | | Deposit date: | 2005-03-14 | | Release date: | 2006-03-14 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Structure of the envelope protein domain III of Omsk hemorrhagic fever virus.
Virology, 351, 2006
|
|
2AXM
 
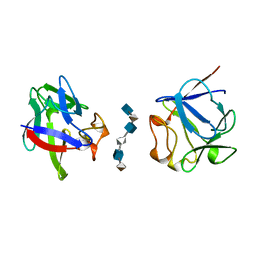 | | HEPARIN-LINKED BIOLOGICALLY-ACTIVE DIMER OF FIBROBLAST GROWTH FACTOR | | Descriptor: | 2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Digabriele, A.D, Lax, I, Chen, D.I, Svahn, C.M, Jaye, M, Schlessinger, J, Hendrickson, W.A. | | Deposit date: | 1997-10-20 | | Release date: | 1998-04-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a heparin-linked biologically active dimer of fibroblast growth factor.
Nature, 393, 1998
|
|
3ROO
 
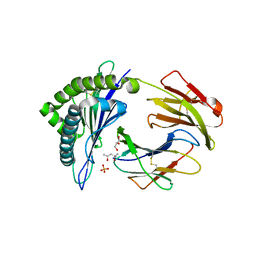 | | Murine class I major histocompatibility complex H-2Kb in complex with immunodominant LCMV-derived gp34-41 peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Madhurantakam, C, Duru, A.D, Leong, C, Sandalova, T, Webb, J.R, Achour, A. | | Deposit date: | 2011-04-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Nitro-tyrosination of the immunodominant LCMV epitope gp34-41 alters both its capacity to stabilize H-2Kb and the molecular surface of the MHC complex, affecting TCR recognition
PLoS ONE, 2012
|
|
3S7B
 
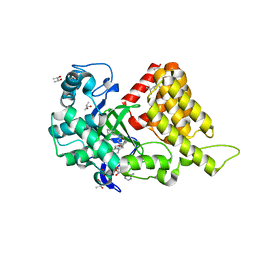 | | Structural Basis of Substrate Methylation and Inhibition of SMYD2 | | Descriptor: | (R,R)-2,3-BUTANEDIOL, N-cyclohexyl-N~3~-[2-(3,4-dichlorophenyl)ethyl]-N-(2-{[2-(5-hydroxy-3-oxo-3,4-dihydro-2H-1,4-benzoxazin-8-yl)ethyl]amino}ethyl)-beta-alaninamide, N-lysine methyltransferase SMYD2, ... | | Authors: | Ferguson, A.D. | | Deposit date: | 2011-05-26 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Methylation and Inhibition of SMYD2.
Structure, 19, 2011
|
|
3TW6
 
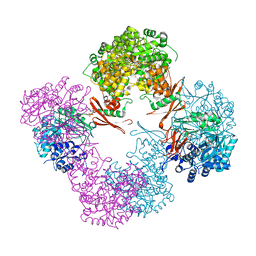 | | Structure of Rhizobium etli pyruvate carboxylase T882A with the allosteric activator, acetyl coenzyme-A | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ... | | Authors: | St Maurice, M, Kumar, S, Lietzan, A.D. | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-19 | | Last modified: | 2011-12-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Interaction between the biotin carboxyl carrier domain and the biotin carboxylase domain in pyruvate carboxylase from Rhizobium etli.
Biochemistry, 50, 2011
|
|
1XYW
 
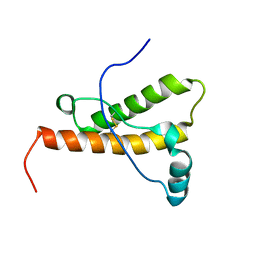 | | elk prion protein | | Descriptor: | Major prion protein | | Authors: | Gossert, A.D, Bonjour, S, Lysek, D.A, Fiorito, F, Wuthrich, K. | | Deposit date: | 2004-11-11 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of elk and of mouse/elk hybrids
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3S7F
 
 | |
3S26
 
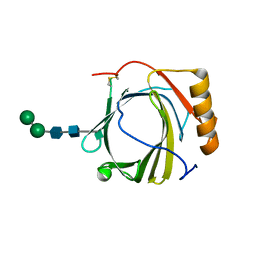 | | Crystal Structure of Murine Siderocalin (Lipocalin 2, 24p3) | | Descriptor: | Neutrophil gelatinase-associated lipocalin, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Correnti, C, Bandaranayake, A.D, Strong, R.K. | | Deposit date: | 2011-05-16 | | Release date: | 2011-09-28 | | Last modified: | 2021-03-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Daedalus: a robust, turnkey platform for rapid production of decigram quantities of active recombinant proteins in human cell lines using novel lentiviral vectors.
Nucleic Acids Res., 39, 2011
|
|
1YK9
 
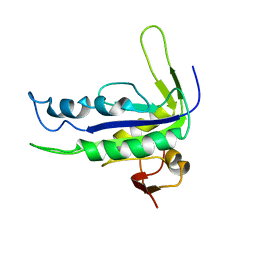 | | Crystal structure of a mutant form of the mycobacterial adenylyl cyclase Rv1625c | | Descriptor: | Adenylate cyclase | | Authors: | Ketkar, A.D, Shenoy, A.R, Ramagopal, U.A, Visweswariah, S.S, Suguna, K, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-01-17 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Structural Basis for the Role of Nucleotide Specifying Residues in Regulating the Oligomerization of the Rv1625c Adenylyl Cyclase from M.tuberculosis
J.Mol.Biol., 356, 2006
|
|
1Y15
 
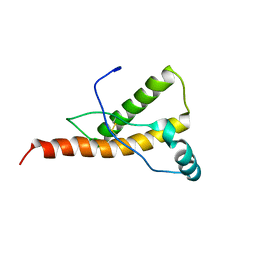 | | Mouse Prion Protein with mutation N174T | | Descriptor: | Major prion protein | | Authors: | Gossert, A.D, Bonjour, S, Lysek, D.A, Fiorito, F, Wuthrich, K. | | Deposit date: | 2004-11-17 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | Prion protein NMR structures of elk and of mouse/elk hybrids
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
3QUL
 
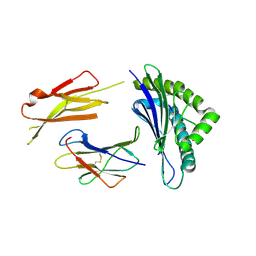 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4S) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
1Y8Y
 
 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | (5-CHLOROPYRAZOLO[1,5-A]PYRIMIDIN-7-YL)-(4-METHANESULFONYLPHENYL)AMINE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.996 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1Y91
 
 | | Crystal structure of human CDK2 complexed with a pyrazolo[1,5-a]pyrimidine inhibitor | | Descriptor: | 4-[5-(TRANS-4-AMINOCYCLOHEXYLAMINO)-3-ISOPROPYLPYRAZOLO[1,5-A]PYRIMIDIN-7-YLAMINO]-N,N-DIMETHYLBENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Williamson, D.S, Parratt, M.J, Torrance, C.J, Bower, J.F, Moore, J.D, Richardson, C.M, Dokurno, P, Cansfield, A.D, Francis, G.L, Hebdon, R.J, Howes, R, Jackson, P.S, Lockie, A.M, Murray, J.B, Nunns, C.L, Powles, J, Robertson, A, Surgenor, A.E. | | Deposit date: | 2004-12-14 | | Release date: | 2005-02-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-guided design of pyrazolo[1,5-a]pyrimidines as inhibitors of human cyclin-dependent kinase 2.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
