3F3A
 
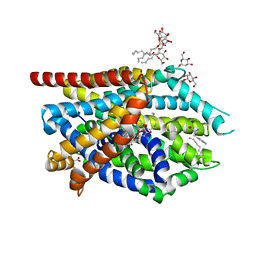 | | Crystal Structure of LeuT bound to L-Tryptophan and Sodium | | Descriptor: | SODIUM ION, TETRADECANE, TRYPTOPHAN, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
3F3C
 
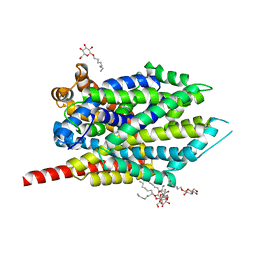 | | Crystal structure of LeuT bound to 4-Fluoro-L-Phenylalanine and sodium | | Descriptor: | 4-FLUORO-L-PHENYLALANINE, SODIUM ION, Transporter, ... | | Authors: | Singh, S.K, Piscitelli, C.L, Yamashita, A, Gouaux, E. | | Deposit date: | 2008-10-30 | | Release date: | 2008-12-23 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A competitive inhibitor traps LeuT in an open-to-out conformation.
Science, 322, 2008
|
|
1J1E
 
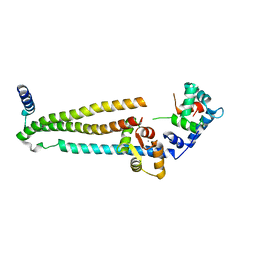 | | Crystal structure of the 52kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
2AE1
 
 | | TROPINONE REDUCTASE-II | | Descriptor: | TROPINONE REDUCTASE-II | | Authors: | Nakajima, K, Yamashita, A, Akama, H, Nakatsu, T, Kato, H, Hashimoto, T, Oda, J, Yamada, Y. | | Deposit date: | 1997-10-27 | | Release date: | 1998-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1J1D
 
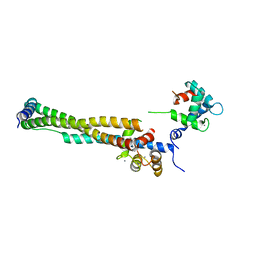 | | Crystal structure of the 46kDa domain of human cardiac troponin in the Ca2+ saturated form | | Descriptor: | CALCIUM ION, Troponin C, Troponin I, ... | | Authors: | Takeda, S, Yamashita, A, Maeda, K, Maeda, Y. | | Deposit date: | 2002-12-03 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structure of the core domain of human cardiac troponin in the Ca2+-saturated form
Nature, 424, 2003
|
|
8KCM
 
 | | MmCPDII-DNA complex containing low-dosage, light induced repaired DNA. | | Descriptor: | Deoxyribodipyrimidine photo-lyase, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, ... | | Authors: | Maestre-Reyna, M, Wang, P.-H, Nango, E, Hosokawa, Y, Saft, M, Furrer, A, Yang, C.-H, Ngura Putu, E.P.G, Wu, W.-J, Emmerich, H.-J, Engilberge, S, Caramello, N, Wranik, M, Glover, H.L, Franz-Badur, S, Wu, H.-Y, Lee, C.-C, Huang, W.-C, Huang, K.-F, Chang, Y.-K, Liao, J.-H, Weng, J.-H, Gad, W, Chang, C.-W, Pang, A.H, Gashi, D, Beale, E, Ozerov, D, Milne, C, Cirelli, C, Bacellar, C, Sugahara, M, Owada, S, Joti, Y, Yamashita, A, Tanaka, R, Tanaka, T, Luo, F.J, Tono, K, Kiontke, S, Spadaccini, R, Royant, A, Yamamoto, J, Iwata, S, Standfuss, J, Essen, L.-O, Bessho, Y, Tsai, M.-D. | | Deposit date: | 2023-08-08 | | Release date: | 2023-11-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Visualizing the DNA repair process by a photolyase at atomic resolution.
Science, 382, 2023
|
|
5X2M
 
 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-glutamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
5X2N
 
 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-alanine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALANINE, CHLORIDE ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
5X2O
 
 | | Crystal structure of the medaka fish taste receptor T1r2a-T1r3 ligand binding domains in complex with L-arginine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ARGININE, CALCIUM ION, ... | | Authors: | Nuemket, N, Yasui, N, Atsumi, N, Yamashita, A. | | Deposit date: | 2017-02-02 | | Release date: | 2017-05-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for perception of diverse chemical substances by T1r taste receptors
Nat Commun, 8, 2017
|
|
1AE1
 
 | | TROPINONE REDUCTASE-I COMPLEX WITH NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TROPINONE REDUCTASE-I | | Authors: | Nakajima, K, Yamashita, A, Akama, H, Nakatsu, T, Kato, H, Hashimoto, T, Oda, J, Yamada, Y. | | Deposit date: | 1997-10-23 | | Release date: | 1998-11-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of two tropinone reductases: different reaction stereospecificities in the same protein fold.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
3WTK
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Thiacloprid | | Descriptor: | Acetylcholine-binding protein, {(2Z)-3-[(6-chloropyridin-3-yl)methyl]-1,3-thiazolidin-2-ylidene}cyanamide | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTH
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTI
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Clothianidin | | Descriptor: | 1-[(2-chloro-1,3-thiazol-5-yl)methyl]-3-methyl-2-nitroguanidine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
3WTN
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Desnitro-imidacloprid | | Descriptor: | (2Z)-1-[(6-chloropyridin-3-yl)methyl]imidazolidin-2-imine, Acetylcholine-binding protein, CADMIUM ION, ... | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
8JZ0
 
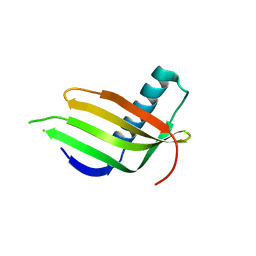 | |
8JZ1
 
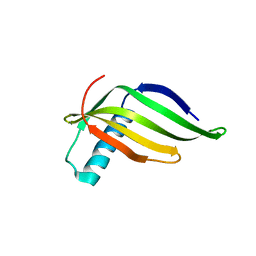 | |
6IR7
 
 | | Green fluorescent protein variant GFPuv with the modification to 6-hydroxynorleucine at the C-terminus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-HYDROXY-L-NORLEUCINE, Green fluorescent protein, ... | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.277 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
6IR6
 
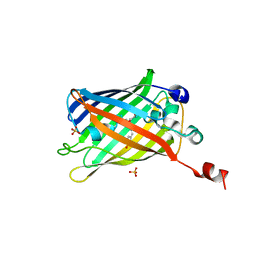 | | Green fluorescent protein variant GFPuv with the native lysine residue at the C-terminus | | Descriptor: | Green fluorescent protein, SULFATE ION | | Authors: | Nakatani, T, Yasui, N, Yamashita, A. | | Deposit date: | 2018-11-12 | | Release date: | 2019-04-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Specific modification at the C-terminal lysine residue of the green fluorescent protein variant, GFPuv, expressed in Escherichia coli.
Sci Rep, 9, 2019
|
|
7CD7
 
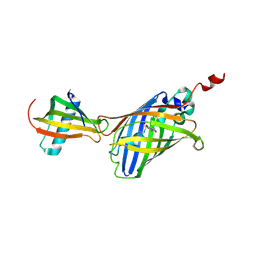 | | GFP-40/GFPuv complex, Form I | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
7CD8
 
 | | GFP-40/GFPuv complex, Form II | | Descriptor: | GFP-40, Green fluorescent protein | | Authors: | Yasui, N, Yamashita, A. | | Deposit date: | 2020-06-18 | | Release date: | 2021-01-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A sweet protein monellin as a non-antibody scaffold for synthetic binding proteins.
J.Biochem., 169, 2021
|
|
3VVI
 
 | |
2ZJU
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Imidacloprid | | Descriptor: | (2E)-1-[(6-chloropyridin-3-yl)methyl]-N-nitroimidazolidin-2-imine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
3WTM
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine-Binding Protein Q55R Mutant Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
2ZJV
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein (Ls-AChBP) Complexed with Clothianidin | | Descriptor: | 1-[(2-chloro-1,3-thiazol-5-yl)methyl]-3-methyl-2-nitroguanidine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Morimoto, T, Matsuda, K. | | Deposit date: | 2008-03-10 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of Lymnaea stagnalis AChBP in complex with neonicotinoid insecticides imidacloprid and clothianidin
Invert.Neurosci., 8, 2008
|
|
3WTL
 
 | | Crystal Structure of Lymnaea stagnalis Acetylcholine Binding Protein Complexed with Nitromethylene Analogue of Imidacloprid | | Descriptor: | 2-chloro-5-{[(2E)-2-(nitromethylidene)imidazolidin-1-yl]methyl}pyridine, Acetylcholine-binding protein | | Authors: | Okajima, T, Ihara, M, Yamashita, A, Oda, T, Matsuda, K. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Studies on an acetylcholine binding protein identify a basic residue in loop G on the beta 1 strand as a new structural determinant of neonicotinoid actions
Mol.Pharmacol., 86, 2014
|
|
