1YE6
 
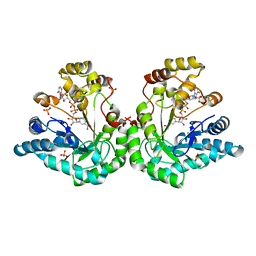 | | Crystal structure of the Lys-274 to Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NADP+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Leitgeb, S, Petschacher, B, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274-->Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD(+) and NADP(+).
Febs Lett., 579, 2005
|
|
3C2R
 
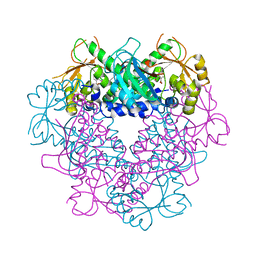 | |
3C2O
 
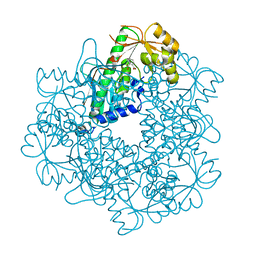 | |
3C2F
 
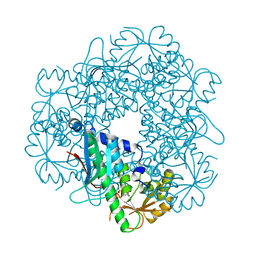 | |
3C2V
 
 | |
3C2E
 
 | |
1PGU
 
 | | YEAST ACTIN INTERACTING PROTEIN 1 (AIP1), Se-Met PROTEIN, MONOCLINIC CRYSTAL FORM | | Descriptor: | Actin interacting protein 1, ZINC ION | | Authors: | Voegtli, W.C, Madrona, A.Y, Wilson, D.K. | | Deposit date: | 2003-05-28 | | Release date: | 2003-07-15 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization.
J.Biol.Chem., 278, 2003
|
|
1PI6
 
 | | YEAST ACTIN INTERACTING PROTEIN 1 (Aip1), ORTHORHOMBIC CRYSTAL FORM | | Descriptor: | Actin interacting protein 1, ZINC ION | | Authors: | Voegtli, W.C, Madrona, A.Y, Wilson, D.K. | | Deposit date: | 2003-05-29 | | Release date: | 2003-07-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of Aip1p, a WD repeat protein that regulates Cofilin-mediated actin depolymerization.
J.Biol.Chem., 278, 2003
|
|
1PZ0
 
 | |
1PZH
 
 | | T.gondii LDH1 ternary complex with NAD and oxalate | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALATE ION, lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1PZG
 
 | |
1PZ1
 
 | |
1PZE
 
 | | T.gondii LDH1 apo form | | Descriptor: | lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
1PYF
 
 | |
1PZF
 
 | | T.gondii LDH1 ternary complex with APAD+ and oxalate | | Descriptor: | 3-ACETYLPYRIDINE ADENINE DINUCLEOTIDE, OXALATE ION, lactate dehydrogenase | | Authors: | Kavanagh, K.L, Elling, R.A, Wilson, D.K. | | Deposit date: | 2003-07-10 | | Release date: | 2004-02-24 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Toxoplasma gondii LDH1: Active-Site Differences from Human Lactate Dehydrogenases and the Structural Basis for Efficient APAD+ Use.
Biochemistry, 43, 2004
|
|
8FH5
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, Aldo-keto reductase family 1 member B1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-001
To Be Published
|
|
8FHC
 
 | | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium | | Descriptor: | BROMIDE ION, CHOLIC ACID, FE (III) ION, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Protein 41 with aldehyde deformylating oxidase activity from Gamma proteobacterium
To Be Published
|
|
8FH6
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001 | | Descriptor: | (8-oxo-7-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-7,8-dihydropyrazino[2,3-d]pyridazin-5-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And Two AT-001
To Be Published
|
|
8FH7
 
 | |
8FH9
 
 | | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007 | | Descriptor: | (4-oxo-3-{[5-(trifluoromethyl)-1,3-benzothiazol-2-yl]methyl}-3,4-dihydrothieno[3,4-d]pyridazin-1-yl)acetic acid, 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member B1, ... | | Authors: | Arenas, R, Wilson, D.K. | | Deposit date: | 2022-12-13 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure Of Aldose Reductase (AKR1B1) Complexed With NADP+ And AT-007
To Be Published
|
|
8FIF
 
 | |
8FHB
 
 | |
8FH8
 
 | |
6D9F
 
 | | Protein 60 with aldehyde deformylating oxidase activity from Kitasatospora setae | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, Putative VlmB homolog, ... | | Authors: | Arenas, R, Wilson, D.K, Mak, W.S, Siegel, J.B. | | Deposit date: | 2018-04-28 | | Release date: | 2019-05-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Discovery, Design, and Structural Characterization of Alkane-Producing Enzymes across the Ferritin-like Superfamily.
Biochemistry, 59, 2020
|
|
5DIC
 
 | |
