1JMK
 
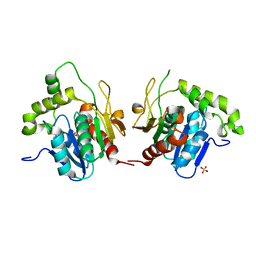 | | Structural Basis for the Cyclization of the Lipopeptide Antibiotic Surfactin by the Thioesterase Domain SrfTE | | Descriptor: | SULFATE ION, Surfactin Synthetase | | Authors: | Bruner, S.D, Weber, T, Kohli, R.M, Schwarzer, D, Marahiel, M.A, Walsh, C.T, Stubbs, M.T. | | Deposit date: | 2001-07-18 | | Release date: | 2002-03-27 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Structural basis for the cyclization of the lipopeptide antibiotic surfactin by the thioesterase domain SrfTE.
Structure, 10, 2002
|
|
1IIR
 
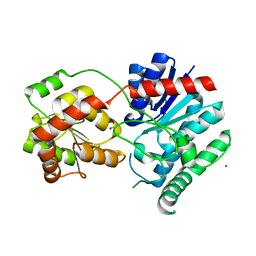 | | Crystal Structure of UDP-glucosyltransferase GtfB | | Descriptor: | MAGNESIUM ION, SULFATE ION, glycosyltransferase GtfB | | Authors: | Mulichak, A.M, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics.
Structure, 9, 2001
|
|
2ROQ
 
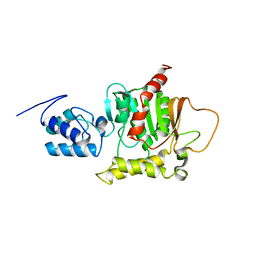 | | Solution Structure of the thiolation-thioesterase di-domain of enterobactin synthetase component F | | Descriptor: | Enterobactin synthetase component F | | Authors: | Frueh, D.P, Arthanari, H, Koglin, A, Vosburg, D.A, Bennett, A.E, Walsh, C.T, Wagner, G. | | Deposit date: | 2008-04-05 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Dynamic thiolation-thioesterase structure of a non-ribosomal peptide synthetase
Nature, 454, 2008
|
|
2RON
 
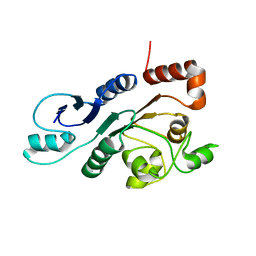 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
2GZR
 
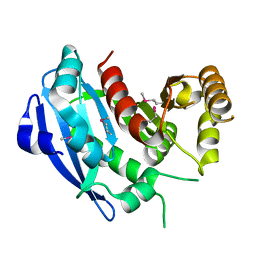 | |
2GZS
 
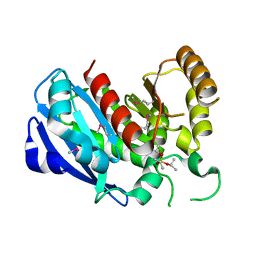 | |
1L5A
 
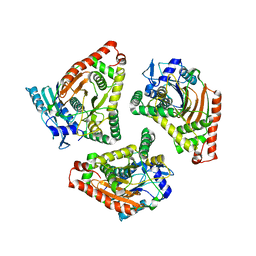 | | Crystal Structure of VibH, an NRPS Condensation Enzyme | | Descriptor: | amide synthase | | Authors: | Keating, T.A, Marshall, C.G, Walsh, C.T, Keating, A.E. | | Deposit date: | 2002-03-06 | | Release date: | 2002-06-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The structure of VibH represents nonribosomal peptide synthetase condensation, cyclization and epimerization domains.
Nat.Struct.Biol., 9, 2002
|
|
2ISK
 
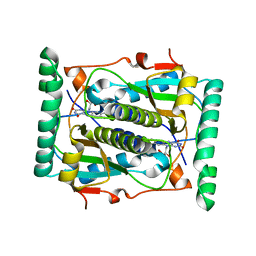 | | BluB bound to flavin anion (charge transfer complex) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, BluB | | Authors: | Larsen, N.A, Taga, M.E, Howard-Jones, A.R, Walsh, C.T, Walker, G.C. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature, 446, 2007
|
|
2ISL
 
 | | BluB bound to reduced flavin (FMNH2) and molecular oxygen. (clear crystal form) | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, BluB, OXYGEN MOLECULE | | Authors: | Larsen, N.A, Taga, M.E, Howard-Jones, A.R, Walsh, C.T, Walker, G.C. | | Deposit date: | 2006-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | BluB cannibalizes flavin to form the lower ligand of vitamin B12.
Nature, 446, 2007
|
|
2K2Q
 
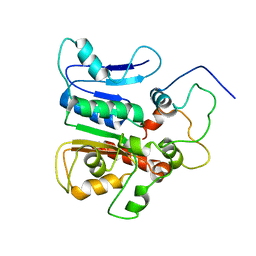 | | complex structure of the external thioesterase of the Surfactin-synthetase with a carrier domain | | Descriptor: | Surfactin synthetase thioesterase subunit, Tyrocidine synthetase 3 (Tyrocidine synthetase III) | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guntert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Dotsch, V. | | Deposit date: | 2008-04-10 | | Release date: | 2008-12-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase.
Nature, 454, 2008
|
|
2MLP
 
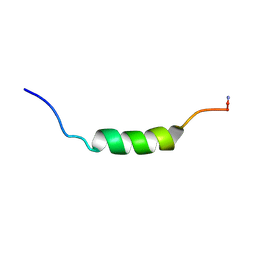 | | MICROCIN LEADER PEPTIDE FROM E. COLI, NMR, 25 STRUCTURES | | Descriptor: | MCBA PROPEPTIDE | | Authors: | Kim, S, Sinha Roy, R, Walsh, C.T, Baleja, J.D. | | Deposit date: | 1998-01-21 | | Release date: | 1998-07-22 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Role of the microcin B17 propeptide in substrate recognition: solution structure and mutational analysis of McbA1-26.
Chem.Biol., 5, 1998
|
|
2LIW
 
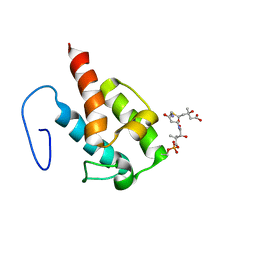 | | NMR structure of HMG-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | 3-HYDROXY-3-METHYL-GLUTARIC ACID, 4'-PHOSPHOPANTETHEINE, CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-21 | | Last modified: | 2025-03-26 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
2LIU
 
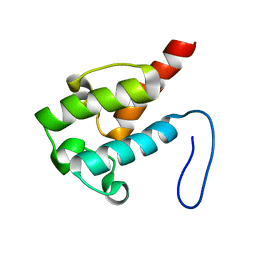 | | NMR structure of holo-ACPI domain from CurA module from Lyngbya majuscula | | Descriptor: | CurA | | Authors: | Busche, A.E, Gottstein, D, Hein, C, Ripin, N, Pader, I, Tufar, P, Eisman, E.B, Gu, L, Walsh, C.T, Loehr, F, Sherman, D.H, Guntert, P, Dotsch, V. | | Deposit date: | 2011-09-01 | | Release date: | 2011-12-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Characterization of Molecular Interactions between ACP and Halogenase Domains in the Curacin A Polyketide Synthase.
Acs Chem.Biol., 7, 2012
|
|
5T81
 
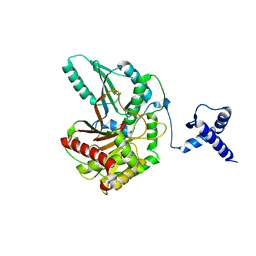 | | Rhombohedral crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB, GLYCEROL | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.604 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5T7Z
 
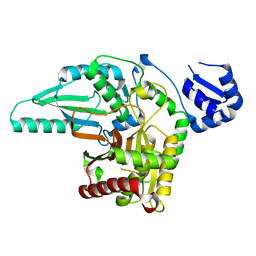 | | Monoclinic crystal form of the EpoB NRPS cyclization-docking bidomain from Sorangium cellulosum | | Descriptor: | EpoB | | Authors: | Dowling, D.P, Kung, Y, Croft, A.K, Taghizadeh, K, Kelly, W.L, Walsh, C.T, Drennan, C.L. | | Deposit date: | 2016-09-06 | | Release date: | 2016-11-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structural elements of an NRPS cyclization domain and its intermodule docking domain.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1IOW
 
 | | COMPLEX OF Y216F D-ALA:D-ALA LIGASE WITH ADP AND A PHOSPHORYL PHOSPHINATE | | Descriptor: | 1(S)-AMINOETHYL-(2-CARBOXYPROPYL)PHOSPHORYL-PHOSPHINIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALA:D-ALA LIGASE, ... | | Authors: | Knox, J.R, Moews, P.C, Fan, C. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | D-alanine:D-alanine ligase: phosphonate and phosphinate intermediates with wild type and the Y216F mutant.
Biochemistry, 36, 1997
|
|
1IOV
 
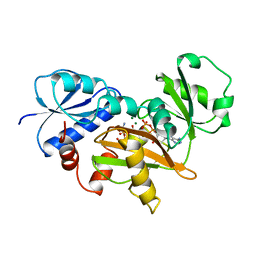 | | COMPLEX OF D-ALA:D-ALA LIGASE WITH ADP AND A PHOSPHORYL PHOSPHONATE | | Descriptor: | 2-[(1-AMINO-ETHYL)-PHOSPHATE-PHOSPHINOYLOXY]-BUTYRIC ACID, ADENOSINE-5'-DIPHOSPHATE, D-ALA:D-ALA LIGASE, ... | | Authors: | Knox, J.R, Moews, P.C, Fan, C. | | Deposit date: | 1996-09-20 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | D-alanine:D-alanine ligase: phosphonate and phosphinate intermediates with wild type and the Y216F mutant.
Biochemistry, 36, 1997
|
|
1OI6
 
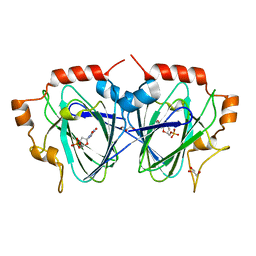 | | Structure determination of the TMP-complex of EvaD | | Descriptor: | GLYCEROL, PCZA361.16, THYMIDINE-5'-PHOSPHATE | | Authors: | Merkel, A.B, Naismith, J.H. | | Deposit date: | 2003-06-09 | | Release date: | 2004-06-03 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Position of a Key Tyrosine in Dtdp-4-Keto-6-Deoxy-D-Glucose-5-Epimerase (Evad) Alters the Substrate Profile for This Rmlc-Like Enzyme
J.Biol.Chem., 279, 2004
|
|
3EPT
 
 | |
1A2N
 
 | | STRUCTURE OF THE C115A MUTANT OF MURA COMPLEXED WITH THE FLUORINATED ANALOG OF THE REACTION TETRAHEDRAL INTERMEDIATE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL-3-FLUORO-2-PHOSPHONOOXY)PROPIONIC ACID | | Authors: | Skarzynski, T. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA
Biochemistry, 37, 1998
|
|
3BMZ
 
 | | Violacein biosynthetic enzyme VioE | | Descriptor: | Putative uncharacterized protein, TETRAETHYLENE GLYCOL, TRIETHYLENE GLYCOL | | Authors: | Ryan, K.S, Drennan, C.L. | | Deposit date: | 2007-12-13 | | Release date: | 2008-01-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | The Violacein Biosynthetic Enzyme VioE Shares a Fold with Lipoprotein Transporter Proteins
J.Biol.Chem., 283, 2008
|
|
3GJA
 
 | | CytC3 | | Descriptor: | ACETATE ION, CytC3 | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
3GJB
 
 | | CytC3 with Fe(II) and alpha-ketoglutarate | | Descriptor: | 2-OXOGLUTARIC ACID, ACETATE ION, CytC3, ... | | Authors: | Wong, C, Drennan, C.L. | | Deposit date: | 2009-03-08 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural analysis of an open active site conformation of nonheme iron halogenase CytC3
J.Am.Chem.Soc., 131, 2009
|
|
4TM3
 
 | |
4TM0
 
 | |
