1C2A
 
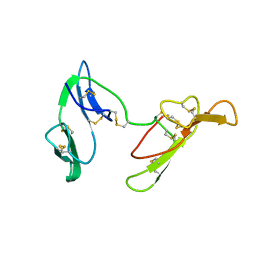 | | CRYSTAL STRUCTURE OF BARLEY BBI | | Descriptor: | BOWMAN-BIRK TRYPSIN INHIBITOR | | Authors: | Song, H.K, Kim, Y.S, Yang, J.K, Moon, J, Lee, J.Y, Suh, S.W. | | Deposit date: | 1999-07-23 | | Release date: | 1999-12-29 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a 16 kDa double-headed Bowman-Birk trypsin inhibitor from barley seeds at 1.9 A resolution.
J.Mol.Biol., 293, 1999
|
|
1C02
 
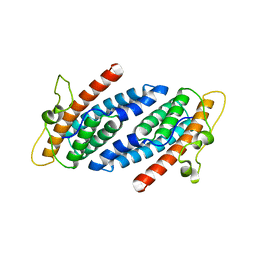 | | CRYSTAL STRUCTURE OF YEAST YPD1P | | Descriptor: | PHOSPHOTRANSFERASE YPD1P | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
1C03
 
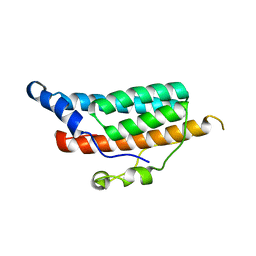 | | CRYSTAL STRUCTURE OF YPD1P (TRICLINIC FORM) | | Descriptor: | HYPOTHETICAL PROTEIN YDL235C | | Authors: | Song, H.K, Lee, J.Y, Lee, M.G, Suh, S.W. | | Deposit date: | 1999-07-14 | | Release date: | 2000-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights into eukaryotic multistep phosphorelay signal transduction revealed by the crystal structure of Ypd1p from Saccharomyces cerevisiae.
J.Mol.Biol., 293, 1999
|
|
6JMZ
 
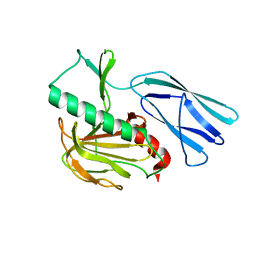 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN0
 
 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN7
 
 | | Structure of H216A mutant closed form peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JMY
 
 | | Structure of wild type closed form of peptidoglycan peptidase | | Descriptor: | CITRIC ACID, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
3KT1
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, GLYCEROL, PKHD-type hydroxylase TPA1, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KT4
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | FE (III) ION, PKHD-type hydroxylase TPA1 | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
3KT7
 
 | | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex | | Descriptor: | 2-OXOGLUTARIC ACID, FE (III) ION, GLYCEROL, ... | | Authors: | Kim, H.S, Kim, H.L, Kim, K.H, Kim, D.J, Lee, S.J, Yoon, J.Y, Yoon, H.J, Lee, H.Y, Park, S.B, Kim, S.-J, Lee, J.Y, Suh, S.W. | | Deposit date: | 2009-11-24 | | Release date: | 2010-01-19 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Crystal structure of Tpa1 from Saccharomyces cerevisiae, a component of the messenger ribonucleoprotein complex
Nucleic Acids Res., 38, 2010
|
|
1BV2
 
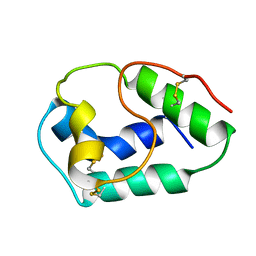 | | LIPID TRANSFER PROTEIN FROM RICE SEEDS, NMR, 14 STRUCTURES | | Descriptor: | NONSPECIFIC LIPID TRANSFER PROTEIN | | Authors: | Poznanski, J, Sodano, P, Suh, S.W, Lee, J.Y, Ptak, M, Vovelle, F. | | Deposit date: | 1998-09-21 | | Release date: | 1999-05-18 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a lipid transfer protein extracted from rice seeds. Comparison with homologous proteins.
Eur.J.Biochem., 259, 1999
|
|
1BXB
 
 | | XYLOSE ISOMERASE FROM THERMUS THERMOPHILUS | | Descriptor: | XYLOSE ISOMERASE | | Authors: | Chang, C, Park, B.C, Lee, D.-S, Suh, S.W. | | Deposit date: | 1998-10-02 | | Release date: | 1999-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of thermostable xylose isomerases from Thermus caldophilus and Thermus thermophilus: possible structural determinants of thermostability.
J.Mol.Biol., 288, 1999
|
|
1DGS
 
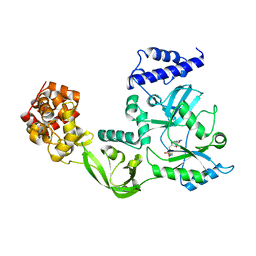 | | CRYSTAL STRUCTURE OF NAD+-DEPENDENT DNA LIGASE FROM T. FILIFORMIS | | Descriptor: | ADENOSINE MONOPHOSPHATE, DNA LIGASE, ZINC ION | | Authors: | Lee, J.Y, Chang, C, Song, H.K, Kwon, S.T, Suh, S.W. | | Deposit date: | 1999-11-25 | | Release date: | 2000-11-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of NAD(+)-dependent DNA ligase: modular architecture and functional implications.
EMBO J., 19, 2000
|
|
3AKJ
 
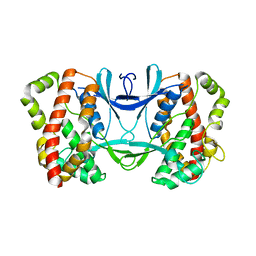 | |
3AKK
 
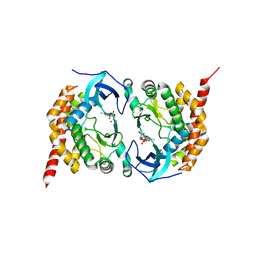 | |
3AKL
 
 | |
1VJS
 
 | |
3CNO
 
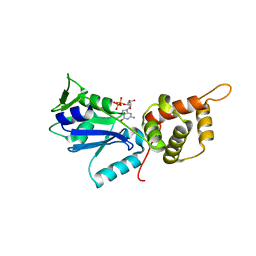 | | GDP-bound structue of TM YlqF | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3CNL
 
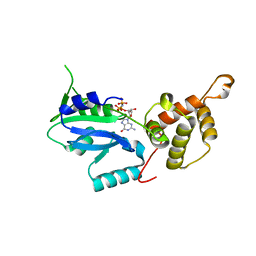 | | Crystal structure of GNP-bound YlqF from T. maritima | | Descriptor: | PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Putative uncharacterized protein | | Authors: | Kim, D.J, Jang, J.Y, Yoon, H.-J, Suh, S.W. | | Deposit date: | 2008-03-26 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of YlqF, a circularly permuted GTPase: Implications for its GTPase activation in 50 S ribosomal subunit assembly
Proteins, 72, 2008
|
|
3F3M
 
 | |
5F8C
 
 | | Rv2258c-unbound | | Descriptor: | GLYCEROL, Methyltransferase | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5F8F
 
 | | Rv2258c-SFG | | Descriptor: | GLYCEROL, Methyltransferase, SINEFUNGIN | | Authors: | Im, H.N, Suh, S.W. | | Deposit date: | 2015-12-09 | | Release date: | 2016-06-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of Rv2258c from Mycobacterium tuberculosis H37Rv, an S-adenosyl-l-methionine-dependent methyltransferase
J.Struct.Biol., 193, 2016
|
|
5GTP
 
 | |
