9CG0
 
 | | Human DJ-1, 30 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K.A, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CFZ
 
 | | Human DJ-1, 20 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography | | Descriptor: | 1-hydroxypropan-2-one, Protein deglycase DJ-1 | | Authors: | Zielinski, K.A, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CFQ
 
 | | Human DJ-1, no mixing, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CMX
 
 | | Human DJ-1, fixed target serial crystallography | | Descriptor: | Parkinson disease protein 7 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-07-15 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CFM
 
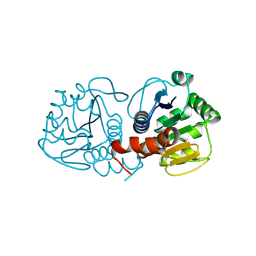 | | Human DJ-1, 5 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography | | Descriptor: | 1-hydroxypropan-2-one, Protein deglycase DJ-1 | | Authors: | Zielinski, K.A, Dolamore, C, Dalton, K, Meisburger, S.P, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-27 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CGB
 
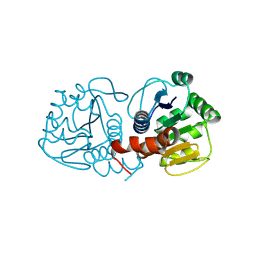 | | Human DJ-1, 5 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | 1-hydroxypropan-2-one, Protein deglycase DJ-1 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CGA
 
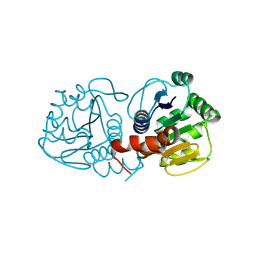 | | Human DJ-1, 3 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | 1-hydroxypropan-2-one, Protein deglycase DJ-1 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CGF
 
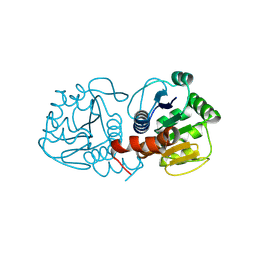 | | Human DJ-1, 20 sec mixing with methylglyoxal, pink beam time-resolved serial crystallography, CrystFEL processed | | Descriptor: | 1-hydroxypropan-2-one, Protein deglycase DJ-1 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-06-28 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
9CMY
 
 | | Human DJ-1, 6.5-18.5 min mixing with methylglyoxal, fixed target serial crystallography | | Descriptor: | Protein deglycase DJ-1 | | Authors: | Zielinski, K, Dolamore, C, Dalton, K, Meisburger, S, Smith, N, Termini, J, Henning, R, Srajer, V, Hekstra, D, Pollack, L, Wilson, M.A. | | Deposit date: | 2024-07-15 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Resolving DJ-1 Glyoxalase Catalysis Using Mix-and-Inject Serial Crystallography at a Synchrotron.
Biorxiv, 2024
|
|
2WZS
 
 | | Structure of the Family GH92 Inverting Mannosidase BT3990 from Bacteroides thetaiotaomicron VPI-5482 in complex with Mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, CALCIUM ION, GLYCEROL, ... | | Authors: | Zhu, Y, Suits, M.D.L, Thompson, A, Chavan, S, Dinev, Z, Dumon, C, Smith, N, Moremen, K.W, Xiang, Y, Siriwardena, A, Williams, S.J, Gilbert, H.J, Davies, G.J. | | Deposit date: | 2009-12-02 | | Release date: | 2009-12-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Mechanistic Insights Into a Ca2+-Dependent Family of A-Mannosidases in a Human Gut Symbiont.
Nat.Chem.Biol., 6, 2010
|
|
2WHJ
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, GLYCEROL, ... | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
2WHL
 
 | | Understanding how diverse mannanases recognise heterogeneous substrates | | Descriptor: | ACETATE ION, BETA-MANNANASE, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Tailford, L.E, Ducros, V.M.A, Flint, J.E, Roberts, S.M, Morland, C, Zechel, D.L, Smith, N, Bjornvad, M.E, Borchert, T.V, Wilson, K.S, Davies, G.J, Gilbert, H.J. | | Deposit date: | 2009-05-05 | | Release date: | 2009-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Understanding How Diverse -Mannanases Recognise Heterogeneous Substrates.
Biochemistry, 48, 2009
|
|
1TT8
 
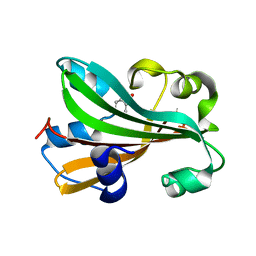 | | CHORISMATE LYASE WITH PRODUCT, 1.0 A RESOLUTION | | Descriptor: | Chorismate-pyruvate lyase, P-HYDROXYBENZOIC ACID | | Authors: | Gallagher, D.T, Mayhew, M, Holden, M.J, Vilker, V, Howard, A. | | Deposit date: | 2004-06-22 | | Release date: | 2004-12-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structural analysis of ligand binding and catalysis in chorismate lyase.
Arch.Biochem.Biophys., 445, 2006
|
|
8OKH
 
 | |
8RYN
 
 | |
8RYQ
 
 | |
8RYO
 
 | |
8RYM
 
 | |
8RYP
 
 | |
9FSK
 
 | | Crystal structure of the HECT domain of Smurf1 | | Descriptor: | E3 ubiquitin-protein ligase SMURF1 | | Authors: | Ostermann, N. | | Deposit date: | 2024-06-21 | | Release date: | 2025-04-16 | | Last modified: | 2025-05-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Therapeutic potential of allosteric HECT E3 ligase inhibition.
Cell, 188, 2025
|
|
9FSH
 
 | |
9FSJ
 
 | |
4X2Q
 
 | | Crystal Structure of Human Aldehyde Dehydrogenase, ALDH1a2 | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Retinal dehydrogenase 2 | | Authors: | Stenkamp, R.E, Le Trong, I, Amory, J.K, Paik, J, Goldstein, A.S. | | Deposit date: | 2014-11-26 | | Release date: | 2015-12-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Synthesis and In Vitro Testing of Bisdichloroacetyldiamine Analogs for Use as a Reversible Male Contraceptive
To Be Published
|
|
1OMO
 
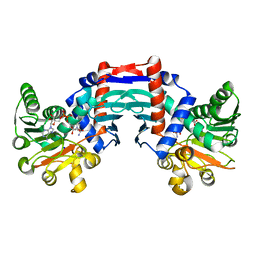 | | alanine dehydrogenase dimer w/bound NAD (archaeal) | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SODIUM ION, alanine dehydrogenase | | Authors: | Gallagher, D.T, Smith, N.N, Holden, M.J, Schroeder, I, Monbouquette, H.G. | | Deposit date: | 2003-02-25 | | Release date: | 2004-07-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of alanine dehydrogenase from Archaeoglobus: active site analysis and relation to bacterial cyclodeaminases and mammalian mu crystallin.
J.Mol.Biol., 342, 2004
|
|
8SMQ
 
 | | Crystal Structure of the N-terminal Domain of the Cryptic Surface Protein (CD630_25440) from Clostridium difficile. | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Kiryukhina, O, Wawrzak, Z, Satchell, K.J.F, Center for Structural Biology of Infectious Diseases (CSBID), Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2023-04-26 | | Release date: | 2023-05-10 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein target highlights in CASP15: Analysis of models by structure providers.
Proteins, 91, 2023
|
|
