1O4U
 
 | |
1O51
 
 | |
1O4T
 
 | |
1O4V
 
 | |
1O4S
 
 | |
2FNO
 
 | |
2FEA
 
 | |
2L6A
 
 | |
1J6U
 
 | |
1ZKG
 
 | |
2AFB
 
 | |
1ZX8
 
 | |
1Z9F
 
 | |
3JXP
 
 | |
6V06
 
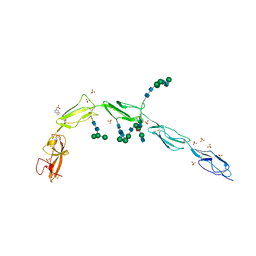 | | Crystal structure of Beta-2 glycoprotein I purified from plasma (pB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-glycoprotein 1, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V09
 
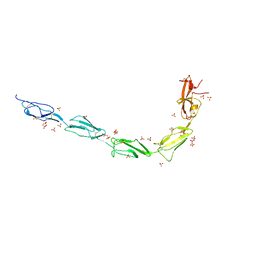 | | Crystal structure of human recombinant Beta-2 glycoprotein I short tag (ST-B2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V08
 
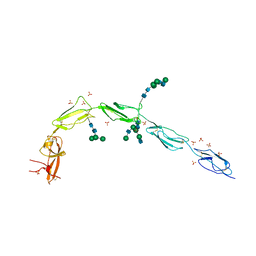 | | Crystal structure of human recombinant Beta-2 glycoprotein I (hrB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
1O5U
 
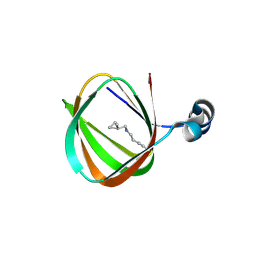 | |
1VJ2
 
 | |
1O4W
 
 | |
1O59
 
 | |
1VJO
 
 | |
1VJ1
 
 | |
1O58
 
 | |
1VKM
 
 | |
