5G5E
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU from mango Mangifera indica L. | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, TAU CLASS GLUTATHIONE S-TRANSFERASE | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-05-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insights Into Ligand Binding to a Glutathione S-Transferase from Mango: Structure, Thermodynamics and Kinetics
Biochimie, 135, 2017
|
|
5G5F
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with reduced glutathione. | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLUTATHIONE, TAU CLASS GLUTATHIONE S-TRANSFERASE | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-05-24 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insights Into Ligand Binding to a Glutathione S-Transferase from Mango: Structure, Thermodynamics and Kinetics
Biochimie, 135, 2017
|
|
2XUW
 
 | | Crystal Structure of Apolaccase from Thermus thermophilus HB27 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, LACCASE | | Authors: | Serrano-Posada, H, Valderrama, B, Rudino-Pinera, E. | | Deposit date: | 2010-10-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.703 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XU9
 
 | | Crystal structure of Laccase from Thermus thermophilus HB27 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Valderrama, B, Rudino-Pinera, E. | | Deposit date: | 2010-10-15 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2XVB
 
 | | Crystal structure of Laccase from Thermus thermophilus HB27 complexed with Hg, crystal of the apoenzyme soaked for 5 min. in 5 mM HgCl2 at 278 K. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, LACCASE, ... | | Authors: | Serrano-Posada, H, Valderrama, B, Rudino-Pinera, E. | | Deposit date: | 2010-10-25 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5KEJ
 
 | | Crystallographic structure of the Tau class glutathione S-transferase MiGSTU in complex with S-hexyl-glutathione | | Descriptor: | DI(HYDROXYETHYL)ETHER, S-HEXYLGLUTATHIONE, Tau class glutathione S-transferase | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A, Hernandez-Paredes, J, Sotelo-Mundo, R. | | Deposit date: | 2016-06-09 | | Release date: | 2017-02-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Insights into ligand binding to a glutathione S-transferase from mango: Structure, thermodynamics and kinetics.
Biochimie, 135, 2017
|
|
4AI7
 
 | | Crystal structure of Laccase from Thermus thermophilus HB27 complexed with Hg, crystal of the apoenzyme soaked for 2 h in 5 mM HgCl2 at 278 K. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, LACCASE-LIKE PROTEIN, MERCURY (II) ION | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2012-02-08 | | Release date: | 2012-02-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAQ
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (75. 0-87.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAH
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (25. 0-37.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAE
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27(0.0- 12.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-21 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAM
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (37. 5-50.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAO
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (50. 0-62.5 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAF
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (12. 5-25.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-22 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAP
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (62. 5-75.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-23 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2YAR
 
 | | X-ray induced reduction of laccase from Thermus thermophilus HB27 (87. 5-100.0 percent dose) | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, COPPER (II) ION, ... | | Authors: | Serrano-Posada, H, Rudino-Pinera, E. | | Deposit date: | 2011-02-24 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray-Induced Catalytic Active-Site Reduction of a Multicopper Oxidase: Structural Insights Into the Proton-Relay Mechanism and O2-Reduction States.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7MIQ
 
 | | Crystal structure of a Glutathione S-transferase class Gtt2 of Vibrio parahaemolyticus (VpGSTT2) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Glutathione S-transferase, SULFATE ION | | Authors: | Valenzuela-Chavira, I, Serrano-Posada, H, Lopez-Zavala, A.A, Garcia-Orozco, K.D, Sotelo-Mundo, R.R. | | Deposit date: | 2021-04-17 | | Release date: | 2021-07-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A Novel Glutathione S -Transferase Gtt2 Class (VpGSTT2) Is Found in the Genome of the AHPND/EMS Vibrio parahaemolyticus Shrimp Pathogen.
Toxins, 13, 2021
|
|
7CE4
 
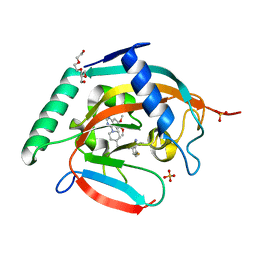 | | Tankyrase2 catalytic domain in complex with K-476 | | Descriptor: | 5-[3-[[1-(6,7-dimethoxyquinazolin-4-yl)piperidin-4-yl]methyl]-2-oxidanylidene-4H-quinazolin-1-yl]-2-fluoranyl-benzenecarbonitrile, Poly [ADP-ribose] polymerase tankyrase-2, SULFATE ION, ... | | Authors: | Takahashi, Y, Suzuki, M, Saito, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-05-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The dual pocket binding novel tankyrase inhibitor K-476 enhances the efficacy of immune checkpoint inhibitor by attracting CD8 + T cells to tumors.
Am J Cancer Res, 11, 2021
|
|
7TVW
 
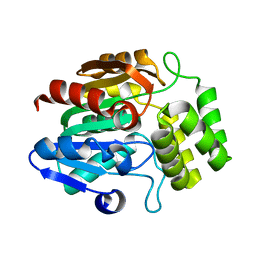 | | Crystal structure of Arabidopsis thaliana DLK2 | | Descriptor: | Alpha/beta-Hydrolases superfamily protein | | Authors: | Burger, M, Chory, J. | | Deposit date: | 2022-02-06 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Crystal structure of Arabidopsis DWARF14-LIKE2 (DLK2) reveals a distinct substrate binding pocket architecture.
Plant Direct, 6, 2022
|
|
7E5V
 
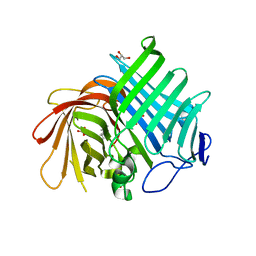 | | Crystal structure of Phm7 in complex with inhibitor | | Descriptor: | Diels-Alderase, GLYCEROL, SULFATE ION, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5U
 
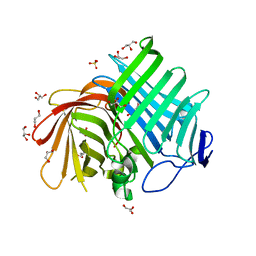 | | Crystal structure of Phm7 | | Descriptor: | CHLORIDE ION, Diels-Alderase, GLYCEROL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7E5T
 
 | | Crystal structure of Fsa2 | | Descriptor: | Diels-Alderase fsa2, ETHANOL, PENTAETHYLENE GLYCOL, ... | | Authors: | Fujiyama, K, Kato, N, Kinugasa, K, Hino, T, Takahashi, S, Nagano, S. | | Deposit date: | 2021-02-20 | | Release date: | 2021-06-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.16977525 Å) | | Cite: | Molecular Basis for Two Stereoselective Diels-Alderases that Produce Decalin Skeletons*.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
7N5K
 
 | | PCNA from Thermococcus gammatolerans: crystal II, collection 1, 1.98 A, 3.84 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5J
 
 | | PCNA from Thermococcus gammatolerans: crystal I, collection 5, 2.82 A, 89.1 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5M
 
 | | PCNA from Thermococcus gammatolerans: crystal III, collection 1, 2.00 A, 1.91 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
7N5I
 
 | | PCNA from Thermococcus gammatolerans: crystal I, collection 1, 1.95 A, 5.22 MGy | | Descriptor: | DNA polymerase sliding clamp, GLYCEROL, SULFATE ION | | Authors: | Marin-Tovar, Y, Rudino-Pinera, E. | | Deposit date: | 2021-06-05 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | PCNA from Thermococcus gammatolerans: A protein involved in chromosomal DNA metabolism intrinsically resistant at high levels of ionizing radiation.
Proteins, 90, 2022
|
|
