5TRN
 
 | | Solution Structure of a DNA Dodecamer with 8-oxoguanine at the 4th position and 5-methylcytosine at the 9th position | | Descriptor: | DNA (5'-D(*CP*GP*CP*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*G)-3') | | Authors: | Hoppins, J.J, Gruber, D.R, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-10-26 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5L06
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd Position | | Descriptor: | DNA (5'-D(*CP*GP*(5CM)P*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-07-26 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5L2G
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th Position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(5CM)P*GP*CP*G)-3') | | Authors: | Miears, H.L, Hoppins, J.J, Gruber, D.R, Kasymov, R.D, Johnson, E.C, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2016-08-01 | | Release date: | 2016-12-21 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
3IF1
 
 | | Crystal structure of 237mAb in complex with a GalNAc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-galactopyranose, Immunoglobulin heavy chain (IgG2a), Immunoglobulin light chain (IgG2a), ... | | Authors: | Brooks, C.L, Evans, S.V, Borisova, S.N. | | Deposit date: | 2009-07-23 | | Release date: | 2010-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Antibody recognition of a unique tumor-specific glycopeptide antigen.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3IET
 
 | | Crystal Structure of 237mAb with antigen | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Immunoglobulin heavy chain (IgG2a), Immunoglobulin light chain (IgG2a), ... | | Authors: | Brooks, C.L, Evans, S.V, Borisova, S.N. | | Deposit date: | 2009-07-23 | | Release date: | 2010-06-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antibody recognition of a unique tumor-specific glycopeptide antigen.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2HF5
 
 | | The structure and function of a novel two-site calcium-binding fragment of calmodulin | | Descriptor: | CALCIUM ION, Calmodulin | | Authors: | Lakowski, T.M, Lee, G.M, Reid, R.E, McIntosh, L.P. | | Deposit date: | 2006-06-23 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Calcium-induced folding of a fragment of calmodulin composed of EF-hands 2 and 3
Protein Sci., 16, 2007
|
|
6CCD
 
 | |
4OYC
 
 | |
5ILS
 
 | | Autoinhibited ETV1 | | Descriptor: | ETS translocation variant 1 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
5ILU
 
 | | Autoinhibited ETV4 | | Descriptor: | ETS translocation variant 4 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.101 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
5ILV
 
 | | Uninhibited ETV5 | | Descriptor: | ETS translocation variant 5 | | Authors: | Whitby, F.G, Currie, S.L. | | Deposit date: | 2016-03-04 | | Release date: | 2017-02-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structured and disordered regions cooperatively mediate DNA-binding autoinhibition of ETS factors ETV1, ETV4 and ETV5.
Nucleic Acids Res., 45, 2017
|
|
6ALT
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALU
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 8-oxoguanine at the 4th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*CP*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
6ALS
 
 | | Solution structure of a DNA dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 4th position | | Descriptor: | DNA (5'-D(*(DC5)P*GP*(DMC)P*(8OG)P*AP*AP*TP*TP*(DMC)P*GP*CP*(DG3))-3') | | Authors: | Gruber, D.R, Shernyukov, A.V, Endutkin, A.V, Bagryanskaya, E.G, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ2
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd and 9th position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5VNT
 
 | |
5UZ1
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 3rd position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*(DMC)P*GP*AP*AP*TP*TP*CP*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
5UZ3
 
 | | Solution Structure of a DNA Dodecamer with 5-methylcytosine at the 9th position and 8-oxoguanine at the 10th position | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(DMC)P*(8OG)P*CP*G)-3') | | Authors: | Gruber, D.R, Hoppins, J.J, Miears, H.L, Endutkin, A.V, Zharkov, D.O, Smirnov, S.L. | | Deposit date: | 2017-02-24 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Oxidative damage to epigenetically methylated sites affects DNA stability, dynamics and enzymatic demethylation.
Nucleic Acids Res., 46, 2018
|
|
2MKY
 
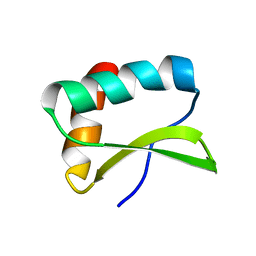 | |
2LF7
 
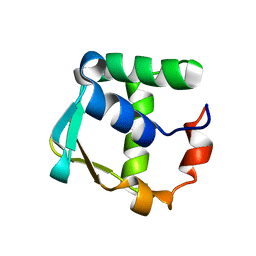 | |
2LF8
 
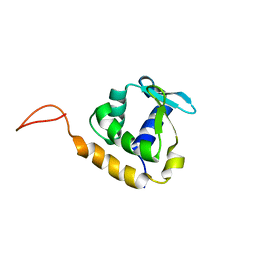 | |
5W3G
 
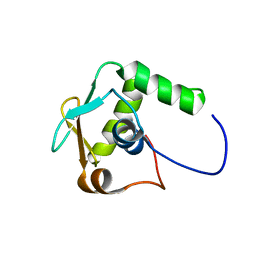 | |
7F4V
 
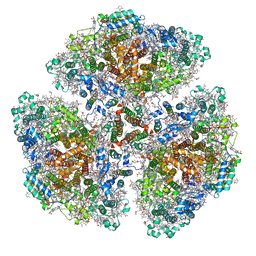 | | Cryo-EM structure of a primordial cyanobacterial photosystem I | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Kato, K, Hamaguchi, T, Nagao, R, Kawakami, K, Yonekura, K, Shen, J.R. | | Deposit date: | 2021-06-21 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.04 Å) | | Cite: | Structural basis for the absence of low-energy chlorophylls responsible for photoprotection from a primitive cyanobacterial PSI
Biorxiv, 2022
|
|
5D5P
 
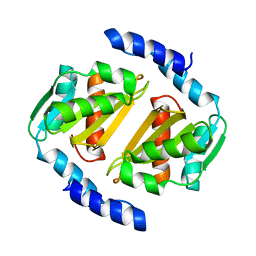 | | HcgB from Methanococcus maripaludis | | Descriptor: | HcgB | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Towards artificial methanogenesis: biosynthesis of the [Fe]-hydrogenase cofactor and characterization of the semi-synthetic hydrogenase.
Faraday Discuss., 198, 2017
|
|
5D5Q
 
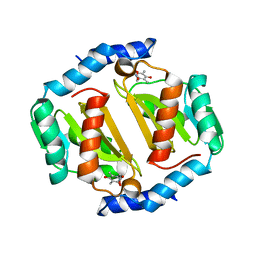 | | HcgB from Methanocaldococcus jannaschii with the pyridinol derived from FeGP cofactor of [Fe]-hydrogenase | | Descriptor: | (4,6-dihydroxy-3,5-dimethylpyridin-2-yl)acetic acid, Uncharacterized protein MJ0488, Guanylyltransferase | | Authors: | Fujishiro, T, Ermler, U, Shima, S. | | Deposit date: | 2015-08-11 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Towards artificial methanogenesis: biosynthesis of the [Fe]-hydrogenase cofactor and characterization of the semi-synthetic hydrogenase.
Faraday Discuss., 198, 2017
|
|
