5J6E
 
 | |
5K58
 
 | |
5K5A
 
 | | Structure of the pNOB8-like ParB N-domain | | Descriptor: | ParB domain protein nuclease | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Method: | X-RAY DIFFRACTION (2.825 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5O
 
 | | Structure of AspA-26mer DNA complex | | Descriptor: | AspA, DNA (26-MER) | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5D
 
 | | Structure of the C2221 form of Pnob8-like ParB-N domain | | Descriptor: | CITRIC ACID, ParB domain protein nuclease | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5R
 
 | | AspA-32mer DNA,crystal form 2 | | Descriptor: | AspA, DNA (32-MER), PHOSPHATE ION | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5K5Q
 
 | |
5K5Z
 
 | | Structure of pnob8 ParA | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ParA | | Authors: | Schumacher, M. | | Deposit date: | 2016-05-24 | | Release date: | 2016-06-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.369 Å) | | Cite: | Structures of archaeal DNA segregation machinery reveal bacterial and eukaryotic linkages.
Science, 349, 2015
|
|
5KHD
 
 | | Structure of 1.75 A BldD C-domain-c-di-GMP complex | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), DNA-binding protein | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-14 | | Release date: | 2016-06-29 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7501 Å) | | Cite: | Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development.
Cell, 158, 2014
|
|
5KBJ
 
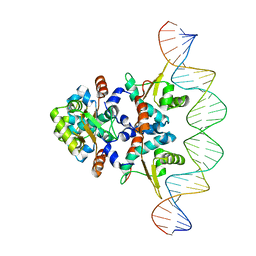 | | Structure of Rep-DNA complex | | Descriptor: | DNA (32-MER), Replication initiator A, N-terminal | | Authors: | Schumacher, M. | | Deposit date: | 2016-06-03 | | Release date: | 2016-06-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Mechanism of staphylococcal multiresistance plasmid replication origin assembly by the RepA protein.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
1EA5
 
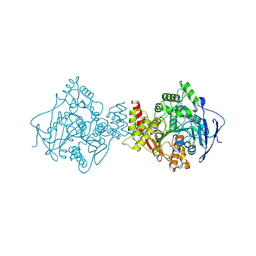 | | NATIVE ACETYLCHOLINESTERASE (E.C. 3.1.1.7) FROM TORPEDO CALIFORNICA at 1.8A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETYLCHOLINESTERASE | | Authors: | Harel, M, Weik, M, Silman, I, Sussman, J.L. | | Deposit date: | 2000-11-06 | | Release date: | 2000-11-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-Ray Structures of Torpedo Californica Acetylcholinesterase Complexed with (+)-Huperzine a and (-)-Huperzine B: Structural Evidence for an Active Site Rearrangement
Biochemistry, 41, 2002
|
|
7QUL
 
 | | Alcohol Dehydrogenase from Thauera aromatica K319A/K320A mutant | | Descriptor: | 1,2-ETHANEDIOL, 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, ZINC ION | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-18 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
7QUY
 
 | | Alcohol Dehydrogenase from Thauera aromatica complexed with NADH | | Descriptor: | 6-hydroxycyclohex-1-ene-1-carbonyl-CoA dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Petchey, M.L, Stark, F, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2022-01-19 | | Release date: | 2022-08-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Advanced Insights into Catalytic and Structural Features of the Zinc-Dependent Alcohol Dehydrogenase from Thauera aromatica.
Chembiochem, 23, 2022
|
|
1MX8
 
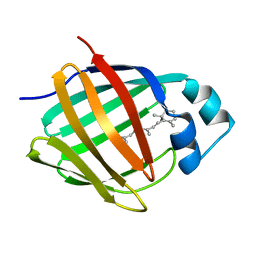 | |
1MX7
 
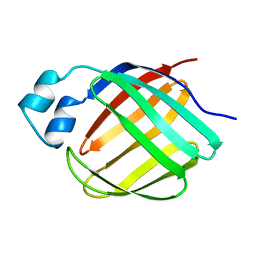 | |
4C4O
 
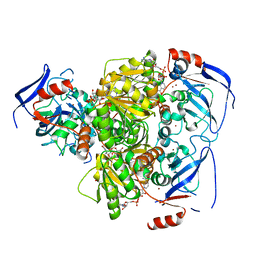 | | Structure of carbonyl reductase CPCR2 from Candida parapsilosis in complex with NADH | | Descriptor: | 1,2-ETHANEDIOL, CARBONYL REDUCTASE CPCR2, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Man, H, Loderer, C, Ansorge-Schumacher, M, Grogan, G. | | Deposit date: | 2013-09-06 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of Nadh-Dependent Carbonyl Reductase (Cpcr2) from Candida Parapsilosis Provides Insight Into Mutations that Improve Catalytic Properties
Chemcatchem, 6, 2014
|
|
2ACE
 
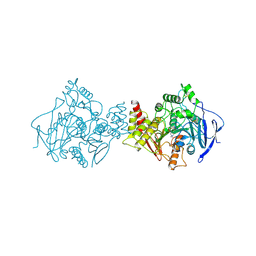 | | NATIVE ACETYLCHOLINESTERASE (E.C. 3.1.1.7) FROM TORPEDO CALIFORNICA | | Descriptor: | ACETYLCHOLINE, ACETYLCHOLINESTERASE | | Authors: | Harel, M, Raves, M.L, Silman, I, Sussman, J.L. | | Deposit date: | 1996-06-23 | | Release date: | 1996-11-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of acetylcholinesterase complexed with the nootropic alkaloid, (-)-huperzine A.
Nat.Struct.Biol., 4, 1997
|
|
3O8N
 
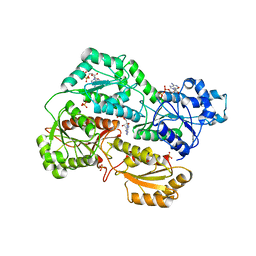 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
3O8O
 
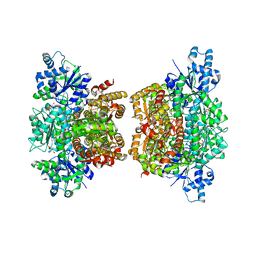 | | Structure of phosphofructokinase from Saccharomyces cerevisiae | | Descriptor: | 2,6-di-O-phosphono-beta-D-fructofuranose, 6-O-phosphono-beta-D-fructofuranose, 6-phosphofructokinase subunit alpha, ... | | Authors: | Banaszak, K, Mechin, I, Kopperschlager, G, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
3O8L
 
 | | Structure of phosphofructokinase from rabbit skeletal muscle | | Descriptor: | 6-phosphofructokinase, muscle type, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Banaszak, K, Chang, S.H, Rypniewski, W. | | Deposit date: | 2010-08-03 | | Release date: | 2011-02-02 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The Crystal Structures of Eukaryotic Phosphofructokinases from Baker's Yeast and Rabbit Skeletal Muscle.
J.Mol.Biol., 407, 2011
|
|
6ELW
 
 | | High resolution structure of selenocysteine containing human GPX4 | | Descriptor: | CHLORIDE ION, Phospholipid hydroperoxide glutathione peroxidase, mitochondrial | | Authors: | Kalms, J, Borchert, A, Kuhn, H, Scheerer, P. | | Deposit date: | 2017-09-29 | | Release date: | 2018-06-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure and functional characterization of selenocysteine-containing glutathione peroxidase 4 suggests an alternative mechanism of peroxide reduction.
Biochim. Biophys. Acta, 1863, 2018
|
|
4E4E
 
 | |
