7PGB
 
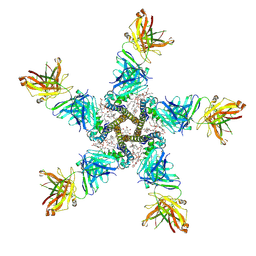 | | NaV_Ae1/Sp1CTD_pore-SAT09 complex | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4-NITROBENZOIC ACID, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PG8
 
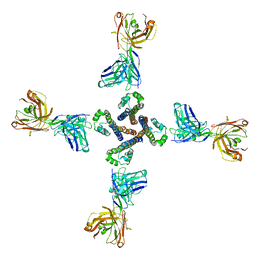 | | NaV_Ae1/Sp1CTD_pore-ANT05 complex | | Descriptor: | ANT05 H12 fab fragment, heavy chain, light chain, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGF
 
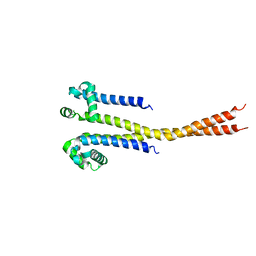 | |
7PGI
 
 | | NaVAb1p (bicelles) | | Descriptor: | ACETATE ION, Ion transport protein, MAGNESIUM ION, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-08 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.638 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGH
 
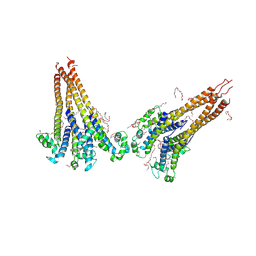 | | NaVAe1/Sp1CTDp (DDM) | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DODECAETHYLENE GLYCOL, DODECANE, ... | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-14 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.194 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7PGG
 
 | | NaVAb1p detergent (DM) | | Descriptor: | 2-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}ethyl heptadecanoate, Ion transport protein | | Authors: | Lolicato, M, Arrigoni, C. | | Deposit date: | 2021-08-13 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Quaternary structure independent folding of voltage-gated ion channel pore domain subunits.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6O0E
 
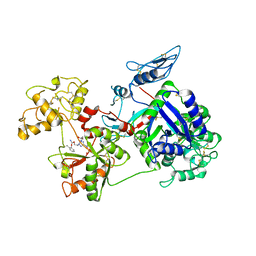 | | Saxiphilin:STX complex, soaking | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Yen, T.J, Lolicato, M, Minor, D.L. | | Deposit date: | 2019-02-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Sci Adv, 5, 2019
|
|
6O0F
 
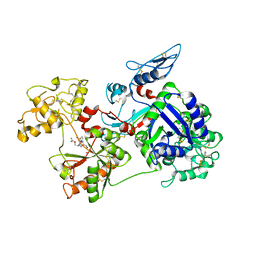 | | Saxiphilin:STX complex, co-crystal | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Yen, T.J, Lolicato, M, Minor, D.L. | | Deposit date: | 2019-02-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Sci Adv, 5, 2019
|
|
6O0D
 
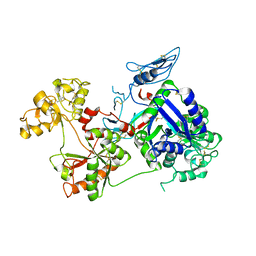 | | Saxiphilin Apo structure | | Descriptor: | Saxiphilin | | Authors: | Yen, T.J, Lolicato, M, Minor, D.L. | | Deposit date: | 2019-02-16 | | Release date: | 2019-07-10 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the saxiphilin:saxitoxin (STX) complex reveals a convergent molecular recognition strategy for paralytic toxins.
Sci Adv, 5, 2019
|
|
6BGJ
 
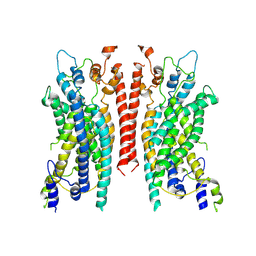 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in LMNG | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, L, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6BGI
 
 | | Cryo-EM structure of the TMEM16A calcium-activated chloride channel in nanodisc | | Descriptor: | Anoctamin-1, CALCIUM ION | | Authors: | Dang, S, Feng, S, Tien, J, Peters, C.J, Bulkley, D, Lolicato, M, Zhao, J, Zuberbuhler, K, Ye, W, Qi, J, Chen, T, Craik, C.S, Jan, Y.N, Minor Jr, D.L, Cheng, Y, Jan, L.Y. | | Deposit date: | 2017-10-28 | | Release date: | 2017-12-27 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-EM structures of the TMEM16A calcium-activated chloride channel.
Nature, 552, 2017
|
|
6V37
 
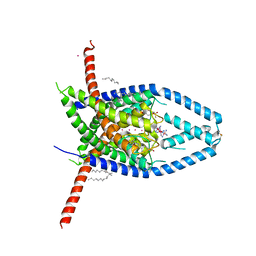 | |
6V3I
 
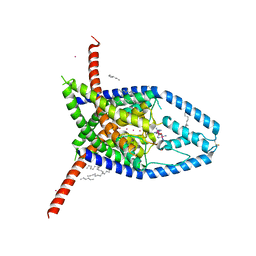 | |
6V36
 
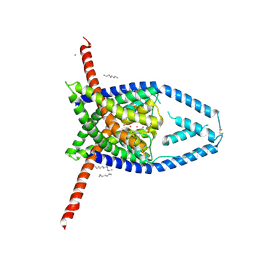 | |
6V3C
 
 | |
4NVP
 
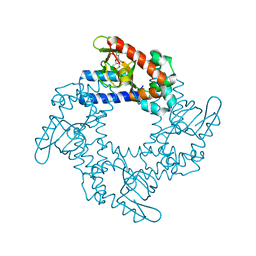 | | Structure of the cyclic nucleotide-binding domain of HCN4 channel complexed with 7-CH-cAMP | | Descriptor: | (2S,4aR,6R,7R,7aS)-6-(4-amino-7H-pyrrolo[2,3-d]pyrimidin-7-yl)tetrahydro-4H-furo[3,2-d][1,3,2]dioxaphosphinine-2,7-diol 2-oxide, Potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 4 | | Authors: | Alfieri, A, Moroni, A. | | Deposit date: | 2013-12-05 | | Release date: | 2014-03-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cyclic Nucleotide Mapping of Hyperpolarization-Activated Cyclic Nucleotide-Gated (HCN) Channels.
Acs Chem.Biol., 9, 2014
|
|
6O3N
 
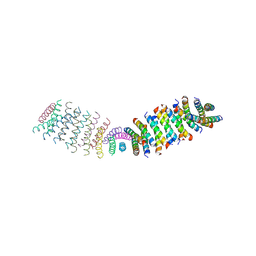 | |
6C4Y
 
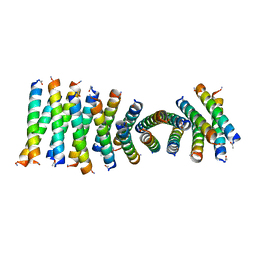 | | Cross-alpha Amyloid-like Structure alphaAmG | | Descriptor: | Cross-alpha Amyloid-like Structure alphaAmG | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6D02
 
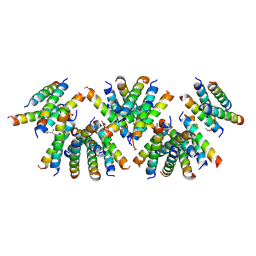 | | Cross-alpha Amyloid-like Structure alphaAmL, 2nd form | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CARBONATE ION, PHOSPHATE ION, ... | | Authors: | Zhang, S.-Q, Liu, L, Degrado, W.F. | | Deposit date: | 2018-04-10 | | Release date: | 2019-02-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C4X
 
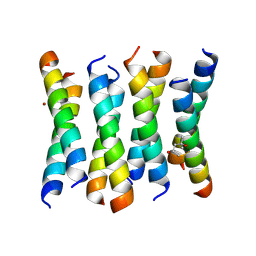 | | Cross-alpha Amyloid-like Structure alphaAmmem | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ZINC ION, cross-alpha amyloid-like membrane peptide alpha-AmMEM | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.55 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C50
 
 | | Cross-alpha Amyloid-like Structure alphaAmS | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmS, FORMIC ACID | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C51
 
 | | Cross-alpha Amyloid-like Structure alphaAmL | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Cross-alpha Amyloid-like Structure alphaAmL, PHOSPHATE ION | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
6C4Z
 
 | |
6C52
 
 | | Cross-alpha Amyloid-like Structure alphaTet | | Descriptor: | Cross-alpha Amyloid-like Structure alphaTet, GLYCEROL | | Authors: | Liu, L, Zhang, S.Q. | | Deposit date: | 2018-01-13 | | Release date: | 2018-08-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Designed peptides that assemble into cross-alpha amyloid-like structures.
Nat. Chem. Biol., 14, 2018
|
|
