7D4M
 
 | | Crystal structure of Tmm from strain HTCC7211 soaked with DMS for 5 min | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.786 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
7D4K
 
 | | Crystal structure of Tmm from Pelagibacter sp. strain HTCC7211 | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Flavin-containing monooxygenase FMO, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2020-09-24 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and Mechanistic Insights Into Dimethylsulfoxide Formation Through Dimethylsulfide Oxidation.
Front Microbiol, 12, 2021
|
|
8ZLZ
 
 | |
8YQP
 
 | | Crystal structure of HylD1 in complex with MEP | | Descriptor: | 2-ethoxycarbonylbenzoic acid, Lipase | | Authors: | Wang, N, Li, C.Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular insights into the catalytic mechanism of a phthalate ester hydrolase.
J Hazard Mater, 476, 2024
|
|
8YQJ
 
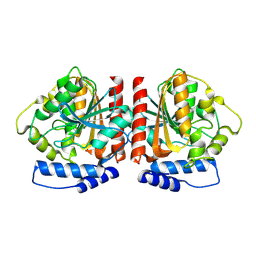 | | Crystal structure of HylD1 | | Descriptor: | Lipase | | Authors: | Wang, N, Li, C.Y. | | Deposit date: | 2024-03-19 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular insights into the catalytic mechanism of a phthalate ester hydrolase.
J Hazard Mater, 476, 2024
|
|
5TOU
 
 | | STRUCTURE OF C-PHYCOCYANIN FROM ARCTIC PSEUDANABAENA SP. LW0831 | | Descriptor: | PHYCOCYANOBILIN, Phycocyanin alpha-1 subunit, Phycocyanin beta-1 subunit | | Authors: | Wang, Q.M, Li, C.Y, Su, H.N, Zhang, Y.Z, Xie, B.B. | | Deposit date: | 2016-10-18 | | Release date: | 2017-03-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural insights into the cold adaptation of the photosynthetic pigment-protein C-phycocyanin from an Arctic cyanobacterium
Biochim. Biophys. Acta, 1858, 2017
|
|
4Q05
 
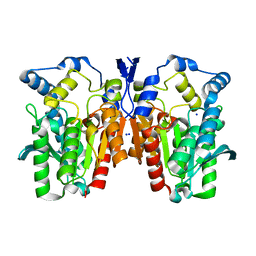 | | Crystal structure of an esterase E25 | | Descriptor: | SODIUM ION, esterase E25 | | Authors: | Li, P.Y, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-03-31 | | Release date: | 2014-06-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for dimerization and catalysis of a novel esterase from the GTSAG motif subfamily of the bacterial hormone-sensitive lipase family
J.Biol.Chem., 289, 2014
|
|
7VVV
 
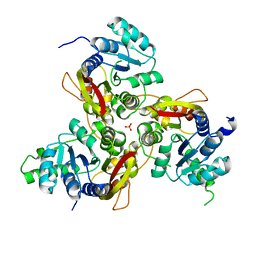 | | Crystal structure of MmtN | | Descriptor: | PHOSPHATE ION, SAM-dependent methyltransferase | | Authors: | Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVW
 
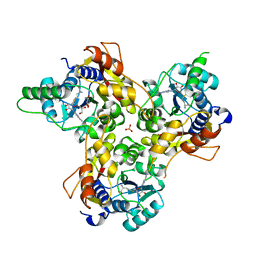 | | MmtN-SAM complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVX
 
 | | MmtN-SAH-Met complex | | Descriptor: | METHIONINE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
8GO3
 
 | |
4Q8K
 
 | | Crystal structure of polysaccharide lyase family 18 aly-SJ02 P-CATD | | Descriptor: | Alginase, CALCIUM ION, SULFATE ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2015-11-25 | | Method: | X-RAY DIFFRACTION (1.646 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
4Q8L
 
 | | Crystal structure of polysacchride lyase family 18 aly-SJ02 r-CATD | | Descriptor: | Alginase, CALCIUM ION | | Authors: | Dong, S, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2014-04-28 | | Release date: | 2014-09-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Molecular insight into the role of the N-terminal extension in the maturation, substrate recognition, and catalysis of a bacterial alginate lyase from polysaccharide lyase family 18.
J.Biol.Chem., 289, 2014
|
|
8HLE
 
 | | Structure of DddY-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, DMSP lyase DddY, ZINC ION | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
6A55
 
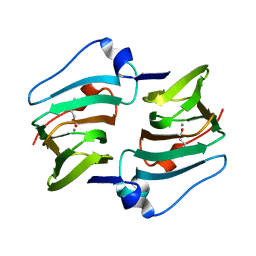 | | Crystal structure of DddK mutant Y122A | | Descriptor: | 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.
Appl. Environ. Microbiol., 85, 2019
|
|
6AK1
 
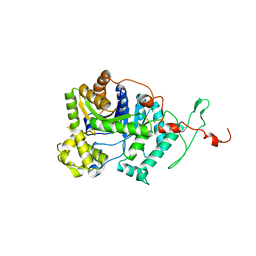 | | Crystal structure of DmoA from Hyphomicrobium sulfonivorans | | Descriptor: | Dimethyl-sulfide monooxygenase | | Authors: | Cao, H.Y, Wang, P, Peng, M, Li, C.Y. | | Deposit date: | 2018-08-28 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.284 Å) | | Cite: | Crystal structure of the dimethylsulfide monooxygenase DmoA from Hyphomicrobium sulfonivorans.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
6A53
 
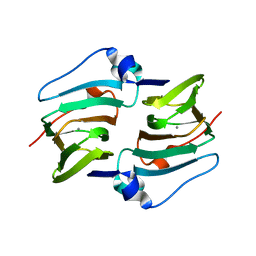 | | Crystal structure of DddK | | Descriptor: | MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.
Appl. Environ. Microbiol., 85, 2019
|
|
6A54
 
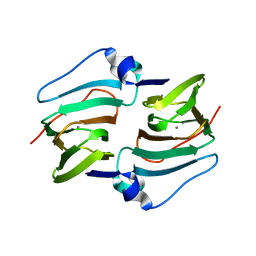 | | Crystal structure of DddK mutant Y64A | | Descriptor: | MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Zhang, Y.Z, Li, C.Y. | | Deposit date: | 2018-06-21 | | Release date: | 2019-02-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure-Function Analysis Indicates that an Active-Site Water Molecule Participates in Dimethylsulfoniopropionate Cleavage by DddK.
Appl. Environ. Microbiol., 85, 2019
|
|
6IJC
 
 | | Structure of MMPA-CoA dehydrogenase from Roseovarius nubinhibens ISM | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Acyl-CoA dehydrogenase family protein | | Authors: | Shao, X, Yuan, Z.L, Cao, H.Y, Wang, P, Li, C.Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
6IHK
 
 | | Structure of MMPA CoA ligase in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, AMP-binding domain protein | | Authors: | Shao, X, Cao, H.Y, Wang, P, Li, C.Y, Zhao, F, Peng, M, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-09-30 | | Release date: | 2019-07-03 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
6IJB
 
 | | Structure of 3-methylmercaptopropionate CoA ligase mutant K523A in complex with AMP and MMPA | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(methylsulfanyl)propanoic acid, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Shao, X, Cao, H.Y, Wang, P, Li, C.Y, Zhao, F, Peng, M, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2018-10-09 | | Release date: | 2019-07-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.111 Å) | | Cite: | Mechanistic insight into 3-methylmercaptopropionate metabolism and kinetical regulation of demethylation pathway in marine dimethylsulfoniopropionate-catabolizing bacteria.
Mol.Microbiol., 111, 2019
|
|
8XZN
 
 | | Crystal structure of folE riboswitch with BH4 | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZW
 
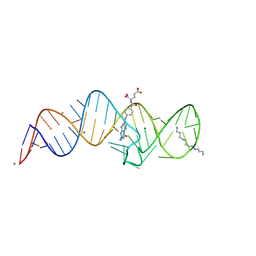 | | Crystal structure of THF-II riboswitch with THF and soaked with Ir | | Descriptor: | (6S)-5,6,7,8-TETRAHYDROFOLATE, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
8XZL
 
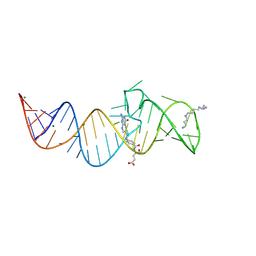 | | Crystal structure of folE riboswitch with DHF | | Descriptor: | DIHYDROFOLIC ACID, MAGNESIUM ION, RNA (53-MER), ... | | Authors: | Li, C.Y, Ren, A.M. | | Deposit date: | 2024-01-21 | | Release date: | 2024-07-24 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure-based characterization and compound identification of the wild-type THF class-II riboswitch.
Nucleic Acids Res., 52, 2024
|
|
