1JCH
 
 | | Crystal Structure of Colicin E3 in Complex with its Immunity Protein | | Descriptor: | CITRIC ACID, COLICIN E3, COLICIN E3 IMMUNITY PROTEIN, ... | | Authors: | Soelaiman, S, Jakes, K, Wu, N, Li, C, Shoham, M. | | Deposit date: | 2001-06-09 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation.
Mol.Cell, 8, 2001
|
|
4JE7
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase in complex with actinonin | | Descriptor: | ACTINONIN, Peptide deformylase 1A, chloroplastic/mitochondrial, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4JE6
 
 | | Crystal structure of a human-like mitochondrial peptide deformylase | | Descriptor: | Peptide deformylase 1A, chloroplastic/mitochondrial, ZINC ION | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2013-02-26 | | Release date: | 2014-02-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Understanding the highly efficient catalysis of prokaryotic peptide deformylases by shedding light on the determinants specifying the low activity of the human counterpart.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1ZY1
 
 | | X-ray structure of peptide deformylase from Arabidopsis thaliana (AtPDF1A) in complex with Met-Ala-Ser | | Descriptor: | Peptide deformylase, mitochondrial, ZINC ION, ... | | Authors: | Fieulaine, S, Juillan-Binard, C, Serero, A, Dardel, F, Giglione, C, Meinnel, T, Ferrer, J.-L. | | Deposit date: | 2005-06-09 | | Release date: | 2005-09-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The crystal structure of mitochondrial (Type 1A) peptide deformylase provides clear guidelines for the design of inhibitors specific for the bacterial forms
J.Biol.Chem., 280, 2005
|
|
1TJ4
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1TJ5
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose and phosphate | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, Sucrose-Phosphatase, ... | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
1U2T
 
 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with sucrose6P | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, sucrose-phosphatase (SPP) | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-07-20 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell
PLANT CELL, 17, 2005
|
|
3PN2
 
 | |
3PN4
 
 | | Crystal structure of Arabidopsis thaliana petide deformylase 1B (AtPDF1B) in complex with actinonin (crystallized in PEG-550-MME) | | Descriptor: | ACTINONIN, Peptide deformylase 1B, chloroplastic, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Trapping conformational States along ligand-binding dynamics of Peptide deformylase: the impact of induced fit on enzyme catalysis.
Plos Biol., 9, 2011
|
|
1U2S
 
 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 in complex with glucose | | Descriptor: | MAGNESIUM ION, alpha-D-glucopyranose, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-07-20 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell
PLANT CELL, 17, 2005
|
|
1S2O
 
 | | X-Ray structure of the sucrose-phosphatase (SPP) from Synechocystis sp. PCC6803 at 1.40 A resolution | | Descriptor: | MAGNESIUM ION, sucrose-phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.L. | | Deposit date: | 2004-01-09 | | Release date: | 2005-02-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
3PN5
 
 | |
3PN3
 
 | | Crystal structure of Arabidopsis thaliana petide deformylase 1B (AtPDF1B) in complex with inhibitor 21 | | Descriptor: | Peptide deformylase 1B, chloroplastic, ZINC ION, ... | | Authors: | Fieulaine, S, Meinnel, T, Giglione, C. | | Deposit date: | 2010-11-18 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Trapping conformational States along ligand-binding dynamics of Peptide deformylase: the impact of induced fit on enzyme catalysis.
Plos Biol., 9, 2011
|
|
3PN6
 
 | |
1TJ3
 
 | | X-Ray structure of the Sucrose-Phosphatase (SPP) from Synechocystis sp. PCC6803 in a closed conformation | | Descriptor: | MAGNESIUM ION, Sucrose-Phosphatase | | Authors: | Fieulaine, S, Lunn, J.E, Borel, F, Ferrer, J.-L. | | Deposit date: | 2004-06-03 | | Release date: | 2005-06-14 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structure of a cyanobacterial sucrose-phosphatase reveals the sugar tongs that release free sucrose in the cell.
Plant Cell, 17, 2005
|
|
2D2V
 
 | |
2B1R
 
 | |
2B1Q
 
 | |
2LM7
 
 | | NMR structure of the C-terminal domain of VP7 in membrane mimicking micelles | | Descriptor: | Outer capsid glycoprotein VP7 | | Authors: | Elaid, S, Libersou, S, Ouldali, M, Morellet, N, Lepault, J, Bouaziz, S. | | Deposit date: | 2011-11-23 | | Release date: | 2012-10-24 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the C-terminal domain of VP7 in membrane mimicking micelles
To be Published
|
|
2KVL
 
 | |
5MTE
 
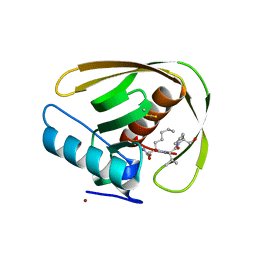 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T in complex with actinonin - crystal form II | | Descriptor: | ACTINONIN, NICKEL (II) ION, Putative uncharacterized protein orf60T, ... | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-11-29 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Peptide deformylases from Vibrio parahaemolyticus phage and bacteria display similar deformylase activity and inhibitor binding clefts.
Biochim. Biophys. Acta, 1866, 2018
|
|
3SVL
 
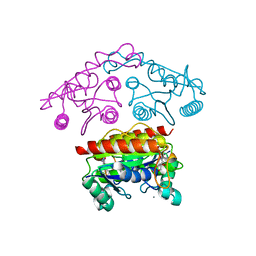 | | Structural basis of the improvement of ChrR - a multi-purpose enzyme | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, protein yieF | | Authors: | Poulain, S, Eswaramoorthy, S, Hienerwadel, R, Bremond, N, Sylvester, M.D, Zhang, Y.B, Van Der Lelie, D, Berthomieu, C, Matin, A.C. | | Deposit date: | 2011-07-12 | | Release date: | 2012-05-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of ChrR-A Quinone Reductase with the Capacity to Reduce Chromate.
Plos One, 7, 2012
|
|
5MTC
 
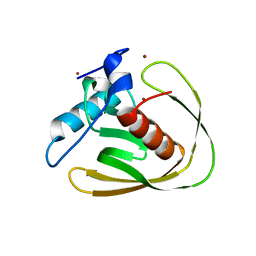 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form I | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein orf60T, ZINC ION | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
5MTD
 
 | | Crystal structure of PDF from the Vibrio parahaemolyticus bacteriophage VP16T - crystal form II | | Descriptor: | NICKEL (II) ION, Putative uncharacterized protein orf60T, TRIETHYLENE GLYCOL, ... | | Authors: | Fieulaine, S, Grzela, R, Giglione, C, Meinnel, T. | | Deposit date: | 2017-01-09 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The C-terminal residue of phage Vp16 PDF, the smallest peptide deformylase, acts as an offset element locking the active conformation.
Sci Rep, 7, 2017
|
|
5A7Q
 
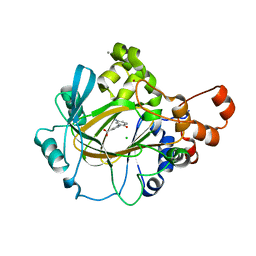 | | Crystal structure of human JMJD2A in complex with compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-azanyl-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Dixon-Clarke, S, MacKenzie, A, Nowak, R, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
