1H4C
 
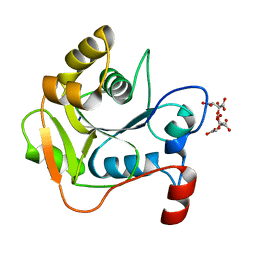 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-26 | | Release date: | 2003-05-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
3ZU2
 
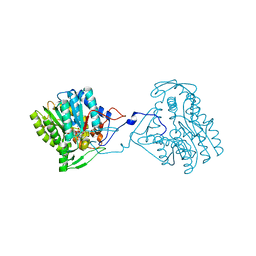 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (SIRAS) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, SODIUM ION | | Authors: | Hirschbeck, M.W, Kuper, J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
5JAM
 
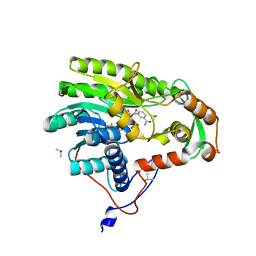 | | Yersinia pestis FabV variant T276V | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
5JAQ
 
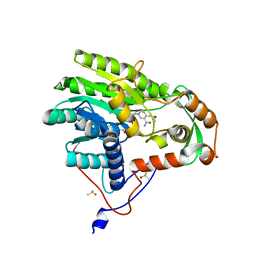 | | Yersinia pestis FabV variant T276C | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-12 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia pestis FabV Enoyl-ACP Reductase.
Biochemistry, 55, 2016
|
|
4F9Z
 
 | | Crystal Structure of human ERp27 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, ACETATE ION, ... | | Authors: | Kober, F.X, Koelmel, W, Kuper, J, Schindelin, H. | | Deposit date: | 2012-05-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of the Protein-Disulfide Isomerase Family Member ERp27 Provides Insights into Its Substrate Binding Capabilities.
J.Biol.Chem., 288, 2013
|
|
6Z4X
 
 | | Structure of the CAK complex form Chaetomium thermophilum bound to ATP-gamma-S | | Descriptor: | CHLORIDE ION, CYCLIN domain-containing protein, MAGNESIUM ION, ... | | Authors: | Peissert, S, Kuper, J, Kisker, C. | | Deposit date: | 2020-05-26 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural basis for CDK7 activation by MAT1 and Cyclin H.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6Z3U
 
 | | Structure of the CAK complex form Chaetomium thermophilum | | Descriptor: | CHLORIDE ION, CYCLIN domain-containing protein, Protein kinase domain-containing protein, ... | | Authors: | Peissert, S, Kuper, J, Kisker, C. | | Deposit date: | 2020-05-22 | | Release date: | 2020-12-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for CDK7 activation by MAT1 and Cyclin H.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5A39
 
 | | Structure of Rad14 in complex with cisplatin containing DNA | | Descriptor: | 5'-D(*DGP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP*GP)-3', 5'-D(*DTP*GP*AP*TP*GP*AP*CP*CP*GP*TP*AP*GP*AP)-3', Cisplatin, ... | | Authors: | Koch, S.C, Kuper, J, Gasteiger, K.L, Wichlein, N, Strasser, R, Eisen, D, Geiger, S, Schneider, S, Kisker, C, Carell, T. | | Deposit date: | 2015-05-28 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into the Recognition of Cisplatin and Aaf-Dg Lesion by Rad14 (Xpa).
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
3IPO
 
 | | Crystal structure of YnjE | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, GLYCEROL, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
3IPP
 
 | | crystal structure of sulfur-free YnjE | | Descriptor: | GLYCEROL, PHOSPHATE ION, Putative thiosulfate sulfurtransferase ynjE, ... | | Authors: | Haenzelmann, P, Kuper, J, Schindelin, H. | | Deposit date: | 2009-08-18 | | Release date: | 2009-12-08 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of YnjE from Escherichia coli, a sulfurtransferase with three rhodanese domains.
Protein Sci., 18, 2009
|
|
1HJL
 
 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
1HJJ
 
 | | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis protein MobA | | Descriptor: | CITRIC ACID, LITHIUM ION, MOLYBDOPTERIN-GUANINE DINUCLEOTIDE BIOSYNTHESIS PROTEIN A | | Authors: | Guse, A, Stevenson, C.E.M, Kuper, J, Buchanan, G, Schwarz, G, Mendel, R.R, Lawson, D.M, Palmer, T. | | Deposit date: | 2003-02-27 | | Release date: | 2003-05-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Biochemical and Structural Analysis of the Molybdenum Cofactor Biosynthesis Protein Moba
J.Biol.Chem., 278, 2003
|
|
3ZU3
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH (MR, cleaved Histag) | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLYCEROL, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZS4
 
 | | CRYSTAL STRUCTURE OF MYCOBACTERIUM TUBERCULOSIS PHOSPHORIBOSYL ISOMERASE WITH BOUND PRFAR | | Descriptor: | PHOSPHORIBOSYL ISOMERASE A, PHOSPHORIC ACID MONO-[5-({[5-CARBAMOYL-3-(5-PHOSPHONOOXY-5-DEOXY-RIBOFURANOSYL)- 3H-IMIDAZOL-4-YLAMINO]-METHYL}-AMINO)-2,3,4-TRIHYDROXY-PENTYL] ESTER | | Authors: | Due, A.V, Kuper, J, Geerlof, A, Wilmanns, M. | | Deposit date: | 2011-06-22 | | Release date: | 2012-07-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Mycobacterium Tuberculosis Phosphoribosyl Isomerase with Bound Prfar
To be Published
|
|
3ZU5
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT173 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(3-amino-2-methylbenzyl)-4-hexylpyridin-2(1H)-one, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
3ZU4
 
 | | Structure of the enoyl-ACP reductase FabV from Yersinia pestis with the cofactor NADH and the 2-pyridone inhibitor PT172 | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 1-(2-CHLOROBENZYL)-4-HEXYLPYRIDIN-2(1H)-ONE, PUTATIVE REDUCTASE YPO4104/Y4119/YP_4011, ... | | Authors: | Hirschbeck, M.W, Kuper, J, Tonge, P.J, Kisker, C. | | Deposit date: | 2011-07-13 | | Release date: | 2012-01-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of the Yersinia Pestis Fabv Enoyl-Acp Reductase and its Interaction with Two 2-Pyridone Inhibitors
Structure, 20, 2012
|
|
5G2O
 
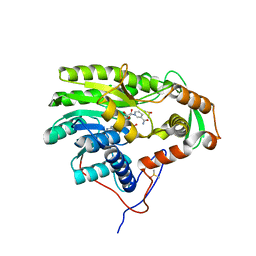 | | Yersinia pestis FabV variant T276A | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, DIMETHYL SULFOXIDE, Enoyl-[acyl-carrier-protein] reductase [NADH] | | Authors: | Pschibul, A, Kuper, J, HIrschbeck, M, Kisker, C. | | Deposit date: | 2016-04-11 | | Release date: | 2016-05-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Selectivity of Pyridone- and Diphenyl Ether-Based Inhibitors for the Yersinia Pestis Fabv Enoyl-Acp Reductase.
Biochemistry, 55, 2016
|
|
4OYU
 
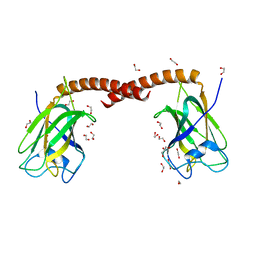 | | Crystal structure of the N-terminal domains of muskelin | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Muskelin | | Authors: | Delto, C, Kuper, J, Schindelin, H. | | Deposit date: | 2014-02-13 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The LisH Motif of Muskelin Is Crucial for Oligomerization and Governs Intracellular Localization.
Structure, 23, 2015
|
|
4PN7
 
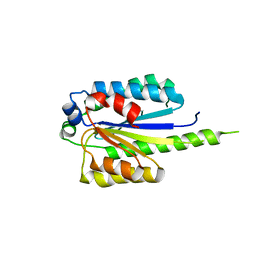 | |
5I4Z
 
 | | Structure of apo OmoMYC | | Descriptor: | CHLORIDE ION, GLYCEROL, Myc proto-oncogene protein, ... | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5I50
 
 | | Structure of OmoMYC bound to double-stranded DNA | | Descriptor: | DNA (5'-D(P*CP*AP*CP*CP*CP*GP*GP*TP*CP*AP*CP*GP*TP*GP*GP*CP*CP*TP*AP*CP*AP*C)-3'), DNA (5'-D(P*GP*TP*GP*TP*AP*GP*GP*CP*CP*AP*CP*GP*TP*GP*AP*CP*CP*GP*GP*GP*TP*G)-3'), Myc proto-oncogene protein | | Authors: | Koelmel, W, Jung, L.A, Kuper, J, Eilers, M, Kisker, C. | | Deposit date: | 2016-02-13 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | OmoMYC blunts promoter invasion by oncogenic MYC to inhibit gene expression characteristic of MYC-dependent tumors.
Oncogene, 36, 2017
|
|
5NUS
 
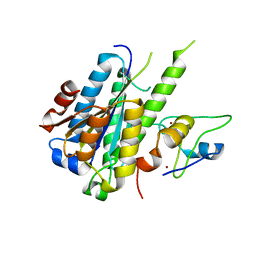 | | Structure of a minimal complex between p44 and p34 from Chaetomium thermophilum | | Descriptor: | ZINC ION, p34, p44 | | Authors: | Koelmel, W, Schoenwetter, E, Kuper, J, Schmitt, D.R, Kisker, C. | | Deposit date: | 2017-05-02 | | Release date: | 2017-10-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
5OBZ
 
 | | low resolution structure of the p34ct/p44ct minimal complex | | Descriptor: | Putative transcription factor, ZINC ION | | Authors: | Schoenwetter, E, Koelmel, W, Schmitt, D.R, Kuper, J, Kisker, C. | | Deposit date: | 2017-06-29 | | Release date: | 2017-10-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | The intricate network between the p34 and p44 subunits is central to the activity of the transcription/DNA repair factor TFIIH.
Nucleic Acids Res., 45, 2017
|
|
5LY9
 
 | | Structure of MITat 1.1 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Variant surface glycoprotein MITAT 1.1, ... | | Authors: | Schaefer, C, Bartossek, T, Jones, N, Kuper, J, Kisker, C, Engstler, M. | | Deposit date: | 2016-09-26 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for the shielding function of the dynamic trypanosome variant surface glycoprotein coat.
Nat Microbiol, 2, 2017
|
|
2W3R
 
 | | Crystal Structure of Xanthine Dehydrogenase (desulfo form) from Rhodobacter capsulatus in complex with hypoxanthine | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dietzel, U, Kuper, J, Leimkuhler, S, Kisker, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism of Substrate and Inhibitor Binding of Rhodobacter Capsulatus Xanthine Dehydrogenase.
J.Biol.Chem., 284, 2009
|
|
