6PGK
 
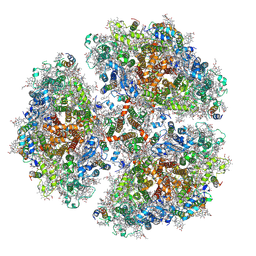 | | Membrane Protein Megahertz Crystallography at the European XFEL, Photosystem I XFEL at 2.9 A | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, BETA-CAROTENE, ... | | Authors: | Fromme, R, Gisriel, C, Fromme, P. | | Deposit date: | 2019-06-24 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Membrane protein megahertz crystallography at the European XFEL.
Nat Commun, 10, 2019
|
|
4K6A
 
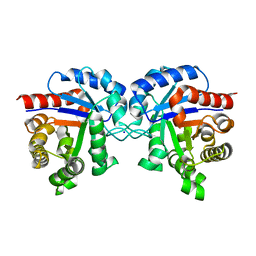 | | Revised Crystal Structure of apo-form of Triosephosphate Isomerase (tpiA) from Escherichia coli at 1.8 Angstrom Resolution. | | Descriptor: | SODIUM ION, Triosephosphate isomerase | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Grimshaw, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-04-15 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural, kinetic and proteomic characterization of acetyl phosphate-dependent bacterial protein acetylation.
PLoS ONE, 9, 2014
|
|
7PYV
 
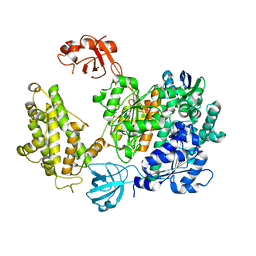 | | Crystal structure of human UBA6 in complex with the ubiquitin-like modifier FAT10 | | Descriptor: | UBD, Ubiquitin-like modifier-activating enzyme 6,Ubiquitin-like modifier-activating enzyme 1,Ubiquitin-like modifier-activating enzyme 6 | | Authors: | Li, S, Truongvan, N, Schindelin, H. | | Deposit date: | 2021-10-11 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
7PVN
 
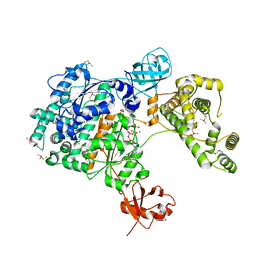 | | Crystal Structure of Human UBA6 in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Truongvan, N, Li, S, Schindelin, H. | | Deposit date: | 2021-10-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
5L6H
 
 | | Uba1 in complex with Ub-ABPA3 covalent adduct | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6I
 
 | | Uba1 in complex with Ub-MLN4924 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
5L6J
 
 | | Uba1 in complex with Ub-MLN7243 covalent adduct | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2016-05-30 | | Release date: | 2017-06-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Dissecting the Specificity of Adenosyl Sulfamate Inhibitors Targeting the Ubiquitin-Activating Enzyme.
Structure, 25, 2017
|
|
4ECM
 
 | | 2.3 Angstrom Crystal Structure of a Glucose-1-phosphate Thymidylyltransferase from Bacillus anthracis in Complex with Thymidine-5-diphospho-alpha-D-glucose and Pyrophosphate | | Descriptor: | 2'DEOXY-THYMIDINE-5'-DIPHOSPHO-ALPHA-D-GLUCOSE, Glucose-1-phosphate thymidylyltransferase, PYROPHOSPHATE 2- | | Authors: | Minasov, G, Kuhn, M, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-03-26 | | Release date: | 2012-04-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme glucose-1-phosphate thymidylyltransferase (RfbA).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4F21
 
 | | Crystal structure of carboxylesterase/phospholipase family protein from Francisella tularensis | | Descriptor: | Carboxylesterase/phospholipase family protein, N-((1R,2S)-2-allyl-4-oxocyclobutyl)-4-methylbenzenesulfonamide, bound form | | Authors: | Filippova, E.V, Minasov, G, Kuhn, M, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J.R, Kiryukhina, O, Becker, D.P, Armoush, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large scale structural rearrangement of a serine hydrolase from Francisella tularensis facilitates catalysis.
J.Biol.Chem., 288, 2013
|
|
6HSO
 
 | | Crystal structure of the ternary complex of GephE-ADP-Glycine receptor derived peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,1'-benzene-1,4-diylbis(1H-pyrrole-2,5-dione), ACETATE ION, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
6HSN
 
 | | Crystal structure of the ternary complex of GephE-ADP-GABA(A) receptor derived peptide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,1'-[ethane-1,2-diylbis(oxyethane-2,1-diyl)]bis(1H-pyrrole-2,5-dione), ACETATE ION, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2018-10-01 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
6FTR
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wiedorn, M.O, Oberthuer, D, Barty, A, Chapman, H.N. | | Deposit date: | 2018-02-23 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.76000106 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
6GTH
 
 | | Serial Femtosecond Crystallography at Megahertz pulse rates | | Descriptor: | (2S,5R)-1-formyl-5-[(sulfooxy)amino]piperidine-2-carboxamide, Beta-lactamase | | Authors: | Wiedorn, M, Oberthuer, D, Werner, N, Schubert, R, White, T.A, Mancuso, A, Perbandt, M, Betzel, C, Barty, A, Chapman, H. | | Deposit date: | 2018-06-18 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Megahertz serial crystallography.
Nat Commun, 9, 2018
|
|
7ZH9
 
 | | Uba1 in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Misra, M, Schindelin, H. | | Deposit date: | 2022-04-05 | | Release date: | 2022-08-31 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structures of UBA6 explain its dual specificity for ubiquitin and FAT10.
Nat Commun, 13, 2022
|
|
8A1I
 
 | |
3RYK
 
 | | 1.63 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose 3,5-epimerase (rfbC) from Bacillus anthracis str. Ames with TDP and PPi bound | | Descriptor: | PYROPHOSPHATE 2-, THYMIDINE-5'-DIPHOSPHATE, dTDP-4-dehydrorhamnose 3,5-epimerase | | Authors: | Halavaty, A.S, Kuhn, M, Minasov, G, Shuvalova, L, Kwon, K, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-11 | | Release date: | 2011-05-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose 3,5-epimerase (RfbC).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
4NNJ
 
 | |
2V58
 
 | | CRYSTAL STRUCTURE OF BIOTIN CARBOXYLASE FROM E.COLI IN COMPLEX WITH POTENT INHIBITOR 1 | | Descriptor: | 6-(2,6-dibromophenyl)pyrido[2,3-d]pyrimidine-2,7-diamine, BIOTIN CARBOXYLASE, CHLORIDE ION | | Authors: | Mochalkin, I, Miller, J.R. | | Deposit date: | 2008-10-02 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Class of Selective Antibacterials Derived from a Protein Kinase Inhibitor Pharmacophore.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2V59
 
 | |
2V5A
 
 | |
6FGC
 
 | | Crystal structure of Gephyrin E domain in complex with Artesunate | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, ... | | Authors: | Kasaragod, V.B, Schindelin, H. | | Deposit date: | 2018-01-10 | | Release date: | 2019-01-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Elucidating the Molecular Basis for Inhibitory Neurotransmission Regulation by Artemisinins.
Neuron, 101, 2019
|
|
6FGD
 
 | |
3NSU
 
 | | A Systematic Screen for Protein-Lipid Interactions in Saccharomyces cerevisiae | | Descriptor: | Phosphatidylinositol 4,5-bisphosphate-binding protein SLM1, SULFATE ION | | Authors: | Gallego, O, Fernandez-Tornero, C, Aguilar-Gurrieri, C, Muller, C, Gavin, A.C. | | Deposit date: | 2010-07-02 | | Release date: | 2010-12-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A systematic screen for protein-lipid interactions in Saccharomyces cerevisiae.
Mol. Syst. Biol., 6, 2010
|
|
6S4Q
 
 | | scdSav(SASK) - Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes | | Descriptor: | GLYCEROL, Streptavidin, {N-(4-{[2-(amino-kappaN)ethyl]sulfamoyl-kappaN}phenyl)-5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentanamide}(chloro)[(1,2,3,4,5-eta)-1,2,3,4,5-pentamethylcyclopentadienyl]iridium(III) | | Authors: | Rebelein, J.G. | | Deposit date: | 2019-06-28 | | Release date: | 2019-09-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Breaking Symmetry: Engineering Single-Chain Dimeric Streptavidin as Host for Artificial Metalloenzymes.
J.Am.Chem.Soc., 141, 2019
|
|
6S50
 
 | |
