4GA3
 
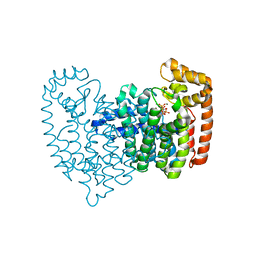 | | Crystal Structure of Human Farnesyl Diphosphate Synthase in Complex with BPH-1260 | | Descriptor: | 1-butyl-3-(2-hydroxy-2,2-diphosphonoethyl)-1H-imidazol-3-ium, Farnesyl pyrophosphate synthase, MAGNESIUM ION, ... | | Authors: | Liu, Y.-L, Zhang, Y, Oldfield, E. | | Deposit date: | 2012-07-24 | | Release date: | 2013-05-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Chemo-Immunotherapeutic Anti-Malarials Targeting Isoprenoid Biosynthesis.
ACS MED.CHEM.LETT., 4, 2013
|
|
7B24
 
 | |
7B1Y
 
 | |
7B23
 
 | |
7B20
 
 | |
7B1V
 
 | |
7B25
 
 | |
6VVA
 
 | |
7MFS
 
 | |
7MQT
 
 | |
7MFN
 
 | |
8B9O
 
 | |
8B9U
 
 | |
7YXU
 
 | | Crystal structure of agonistic antibody 1618 fab domain bound to human 4-1BB. | | Descriptor: | MANGANESE (II) ION, Tumor necrosis factor receptor superfamily member 9, heavy chain of Fab, ... | | Authors: | Hakansson, M, Rose, N, Petersson, J, Enell Smith, K, Thorolfsson, M, von Schantz, L. | | Deposit date: | 2022-02-16 | | Release date: | 2023-01-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The Bispecific Tumor Antigen-Conditional 4-1BB x 5T4 Agonist, ALG.APV-527, Mediates Strong T-Cell Activation and Potent Antitumor Activity in Preclinical Studies.
Mol.Cancer Ther., 22, 2023
|
|
3PCD
 
 | | PROTOCATECHUATE 3,4-DIOXYGENASE Y447H MUTANT | | Descriptor: | BETA-MERCAPTOETHANOL, CARBONATE ION, FE (III) ION, ... | | Authors: | Orville, A.M, Lipscomb, J.D, Ohlendorf, D.H. | | Deposit date: | 1997-11-24 | | Release date: | 1998-05-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The axial tyrosinate Fe3+ ligand in protocatechuate 3,4-dioxygenase influences substrate binding and product release: evidence for new reaction cycle intermediates.
Biochemistry, 37, 1998
|
|
