3LF6
 
 | | Crystal structure of HIV epitope-scaffold 4E10_1XIZA_S0_001_N | | Descriptor: | PHOSPHATE ION, Putative phosphotransferase system | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-15 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
3LG7
 
 | | Crystal structure of HIV epitope-scaffold 4E10_S0_1EZ3A_002_C | | Descriptor: | 4E10_S0_1EZ3A_002_C (T246), SULFATE ION | | Authors: | Holmes, M.A. | | Deposit date: | 2010-01-19 | | Release date: | 2010-09-22 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Computational Design of Epitope-Scaffolds Allows Induction of Antibodies Specific for a Poorly Immunogenic HIV Vaccine Epitope.
Structure, 18, 2010
|
|
1KTL
 
 | | The human non-classical major histocompatibility complex molecule HLA-E | | Descriptor: | BETA-2-MICROGLOBULIN, HLA CLASS I HISTOCOMPATIBILITY ANTIGEN, ALPHA CHAIN, ... | | Authors: | Holmes, M.A, Strong, R.K. | | Deposit date: | 2002-01-16 | | Release date: | 2003-02-25 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | HLA-E allelic variants: Correlating differential expression, peptide affinities, crystal structures and thermal stabilities
J.Biol.Chem., 278, 2003
|
|
4L8I
 
 | | Crystal structure of RSV epitope scaffold FFL_005 | | Descriptor: | RSV epitope scaffold FFL_005 | | Authors: | Jardine, J, Correnti, C, Holmes, M.A, Strong, R.K, Schief, W.R. | | Deposit date: | 2013-06-17 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Proof of principle for epitope-focused vaccine design.
Nature, 507, 2014
|
|
3OMZ
 
 | |
1TMN
 
 | |
1LNE
 
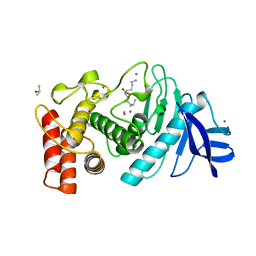 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CADMIUM ION, CALCIUM ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
4TMN
 
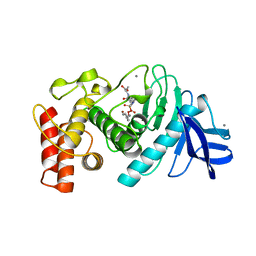 | | SLOW-AND FAST-BINDING INHIBITORS OF THERMOLYSIN DISPLAY DIFFERENT MODES OF BINDING. CRYSTALLOGRAPHIC ANALYSIS OF EXTENDED PHOSPHONAMIDATE TRANSITION-STATE ANALOGUES | | Descriptor: | CALCIUM ION, N-[(S)-[(1R)-1-{[(benzyloxy)carbonyl]amino}-2-phenylethyl](hydroxy)phosphoryl]-L-leucyl-L-alanine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
3TMN
 
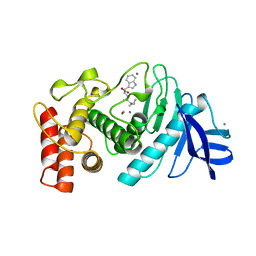 | |
2TMN
 
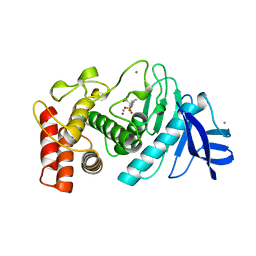 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | Descriptor: | CALCIUM ION, N~2~-phosphono-L-leucinamide, Thermolysin, ... | | Authors: | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
6TMN
 
 | | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond | | Descriptor: | CALCIUM ION, N-[(2R,4S)-4-hydroxy-2-(2-methylpropyl)-4-oxido-7-oxo-9-phenyl-3,8-dioxa-6-aza-4-phosphanonan-1-oyl]-L-leucine, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Holden, H.M, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond.
Science, 235, 1987
|
|
8TLN
 
 | |
1TLP
 
 | | CRYSTALLOGRAPHIC STRUCTURAL ANALYSIS OF PHOSPHORAMIDATES AS INHIBITORS AND TRANSITION-STATE ANALOGS OF THERMOLYSIN | | Descriptor: | CALCIUM ION, N-ALPHA-L-RHAMNOPYRANOSYLOXY(HYDROXYPHOSPHINYL)-L-LEUCYL-L-TRYPTOPHAN, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Monzingo, A.F, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystallographic structural analysis of phosphoramidates as inhibitors and transition-state analogs of thermolysin.
Eur.J.Biochem., 157, 1986
|
|
9EIE
 
 | |
1LNC
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1LNA
 
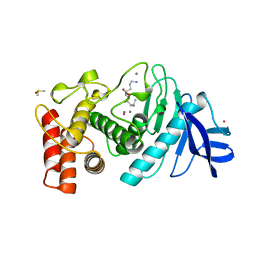 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, COBALT (II) ION, DIMETHYL SULFOXIDE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
1LNF
 
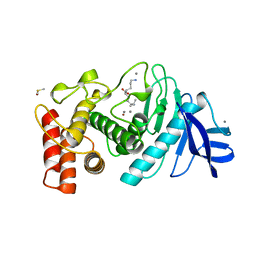 | |
1LNB
 
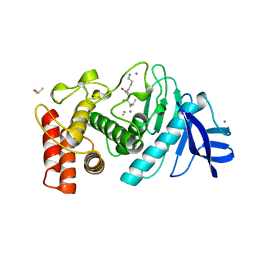 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, FE (III) ION, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
5TMN
 
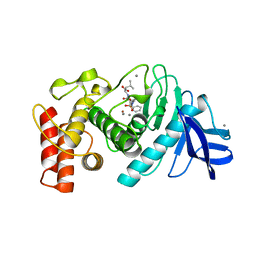 | | Slow-and fast-binding inhibitors of thermolysin display different modes of binding. crystallographic analysis of extended phosphonamidate transition-state analogues | | Descriptor: | CALCIUM ION, N-[(S)-({[(benzyloxy)carbonyl]amino}methyl)(hydroxy)phosphoryl]-L-leucyl-L-leucine, THERMOLYSIN, ... | | Authors: | Holden, H.M, Tronrud, D.E, Monzingo, A.F, Weaver, L.H, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Slow- and fast-binding inhibitors of thermolysin display different modes of binding: crystallographic analysis of extended phosphonamidate transition-state analogues.
Biochemistry, 26, 1987
|
|
1LND
 
 | | A STRUCTURAL ANALYSIS OF METAL SUBSTITUTIONS IN THERMOLYSIN | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, LYSINE, ... | | Authors: | Holland, D.R, Hausrath, A.C, Juers, D, Matthews, B.W. | | Deposit date: | 1994-05-13 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural analysis of zinc substitutions in the active site of thermolysin.
Protein Sci., 4, 1995
|
|
3U9P
 
 | | Crystal Structure of Murine Siderocalin in Complex with an Fab Fragment | | Descriptor: | Monoclonal Fab Fragment Heavy Chain, Monoclonal Fab Fragment Light Chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Correnti, C, Strong, R.K. | | Deposit date: | 2011-10-19 | | Release date: | 2013-05-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Siderocalin/Lcn2/NGAL/24p3 does not drive apoptosis through gentisic acid mediated iron withdrawal in hematopoietic cell lines.
Plos One, 7, 2012
|
|
3U0D
 
 | |
4JLR
 
 | | Crystal structure of a designed Respiratory Syncytial Virus Immunogen in complex with Motavizumab | | Descriptor: | Motavizumab Fab heavy chain, Motavizumab Fab light chain, PENTAETHYLENE GLYCOL, ... | | Authors: | Rupert, P.B, Correia, B, Schief, W, Strong, R.K. | | Deposit date: | 2013-03-12 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Proof of principle for epitope-focused vaccine design.
Nature, 507, 2014
|
|
1XN4
 
 | |
1X9G
 
 | |
