7XN1
 
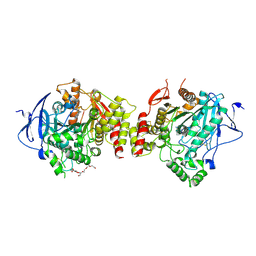 | | Crystal structure of human acetylcholinesterase in complex with tacrine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, Acetylcholinesterase, ... | | Authors: | Dileep, K.V, Ihara, K, Mishima-Tsumagari, C, Kukimoto-Niino, M, Yonemochi, M, Hanada, K, Shirouzu, M, Zhang, K.Y.J. | | Deposit date: | 2022-04-27 | | Release date: | 2023-03-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Crystal structure of human acetylcholinesterase in complex with tacrine: Implications for drug discovery
Int.J.Biol.Macromol., 210, 2022
|
|
6KC6
 
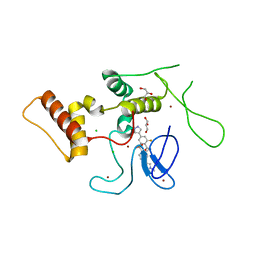 | | HOIP-HOIPIN8 complex | | Descriptor: | 2-[3-[2,6-bis(fluoranyl)-4-(1~{H}-pyrazol-4-yl)phenyl]-3-oxidanylidene-propyl]-4-(1-methylpyrazol-4-yl)benzoic acid, CHLORIDE ION, E3 ubiquitin-protein ligase RNF31, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-06-27 | | Release date: | 2020-04-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.123 Å) | | Cite: | Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses.
Commun Biol, 3, 2020
|
|
8JG5
 
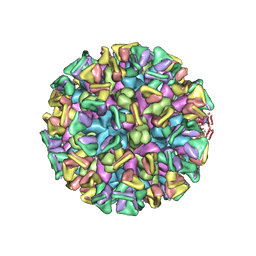 | | Cryo-EM structure of the GI.4 Chiba VLP complexed with the CV-1A1 Fv-clasp | | Descriptor: | VH,SARAH, VL,SARAH, VP1 | | Authors: | Hosaka, T, Katsura, K, Kimura-Someya, T, Someya, Y, Shirouzu, M. | | Deposit date: | 2023-05-19 | | Release date: | 2024-04-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Structural analyses of the GI.4 norovirus by cryo-electron microscopy and X-ray crystallography revealing binding sites for human monoclonal antibodies.
J.Virol., 98, 2024
|
|
8I5L
 
 | |
3H3T
 
 | | Crystal structure of the CERT START domain in complex with HPA-16 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]hexadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3R
 
 | | Crystal structure of the CERT START domain in complex with HPA-14 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tetradecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3Q
 
 | | Crystal structure of the CERT START domain in complex with HPA-13 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]tridecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
3H3S
 
 | | Crystal structure of the CERT START domain in complex with HPA-15 | | Descriptor: | Goodpasture antigen binding protein, N-[(1R,3R)-3-hydroxy-1-(hydroxymethyl)-3-phenylpropyl]pentadecanamide | | Authors: | Kudo, N, Wakatsuki, S, Kato, R. | | Deposit date: | 2009-04-17 | | Release date: | 2010-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Crystal structures of the CERT START domain with inhibitors provide insights into the mechanism of ceramide transfer.
J.Mol.Biol., 396, 2010
|
|
6KC5
 
 | | HOIP-HOIPIN1 complex | | Descriptor: | 2-[3-(2-methoxyphenyl)-3-oxidanylidene-propyl]benzoic acid, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, E3 ubiquitin-protein ligase RNF31, ... | | Authors: | Sato, Y, Fukai, S. | | Deposit date: | 2019-06-27 | | Release date: | 2020-04-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.543 Å) | | Cite: | Molecular bases for HOIPINs-mediated inhibition of LUBAC and innate immune responses.
Commun Biol, 3, 2020
|
|
6JPP
 
 | |
7EIC
 
 | |
7EID
 
 | |
7EIE
 
 | | Crystal structure of YEATS2 YEATS domain | | Descriptor: | GLYCEROL, YEATS domain-containing protein 2 | | Authors: | Kikuchi, M, Umehara, T. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Elucidation of binding preferences of YEATS domains to site-specific acetylated nucleosome core particles.
J.Biol.Chem., 298, 2022
|
|
7X91
 
 | |
7X8W
 
 | |
7X8Y
 
 | |
7X8Z
 
 | |
7X90
 
 | |
7X92
 
 | |
7X93
 
 | |
7X95
 
 | |
7X94
 
 | |
7X96
 
 | |
7VP0
 
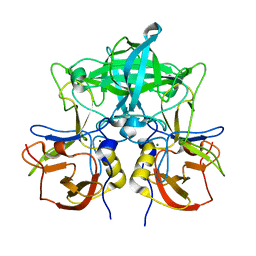 | | Crystal structure of P domain from norovirus GI.9 capsid protein. | | Descriptor: | MAGNESIUM ION, VP1 | | Authors: | Katsura, K, Sakai, N, Hasegawa, K, Kimura-Someya, T, Shirouzu, M. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lewis fucose is a key moiety for the recognition of histo-blood group antigens by GI.9 norovirus, as revealed by structural analysis.
Febs Open Bio, 12, 2022
|
|
7Y6L
 
 | |
