1BDJ
 
 | | COMPLEX STRUCTURE OF HPT DOMAIN AND CHEY | | Descriptor: | AEROBIC RESPIRATION CONTROL SENSOR PROTEIN ARCB, CHEY, SULFATE ION | | Authors: | Kato, M, Mizuno, T, Shimizu, T, Hakoshima, T. | | Deposit date: | 1998-05-10 | | Release date: | 1999-05-11 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Structure of the histidine-containing phosphotransfer (HPt) domain of the anaerobic sensor protein ArcB complexed with the chemotaxis response regulator CheY.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1DJ6
 
 | | COMPLEX OF A Z-DNA HEXAMER, D(CG)3, WITH SYNTHETIC POLYAMINE AT ROOM TEMPERATURE | | Descriptor: | 5'-D(*CP*GP*CP*GP*CP*G)-3', MAGNESIUM ION, N,N'-BIS(2-AMINOETHYL)-1,2-ETHANEDIAMINE | | Authors: | Ohishi, H, Tomita, K.-i, Nakanishi, I, Ohtsuchi, M, Hakoshima, T, Rich, A. | | Deposit date: | 1999-12-01 | | Release date: | 1999-12-18 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | The crystal structure of N1-[2-(2-amino-ethylamino)-ethyl]-ethane-1,2-diamine (polyamines) binding to the minor groove of d(CGCGCG)2, hexamer at room temperature
FEBS Lett., 523, 2002
|
|
2H9V
 
 | | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y27632 | | Descriptor: | (R)-TRANS-4-(1-AMINOETHYL)-N-(4-PYRIDYL) CYCLOHEXANECARBOXAMIDE, Rho-associated protein kinase 2 | | Authors: | Yamaguchi, H, Miwa, Y, Kasa, M, Kitano, K, Amano, M, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2006-06-12 | | Release date: | 2006-12-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for induced-fit binding of Rho-kinase to the inhibitor Y-27632
J.Biochem.(Tokyo), 140, 2006
|
|
1A2B
 
 | | HUMAN RHOA COMPLEXED WITH GTP ANALOGUE | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, TRANSFORMING PROTEIN RHOA | | Authors: | Ihara, K, Muraguchi, S, Kato, M, Shimizu, T, Shirakawa, M, Kuroda, S, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 1997-12-26 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue.
J.Biol.Chem., 273, 1998
|
|
1A0A
 
 | | PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*TP*AP*GP*TP*CP*CP*CP*AP*CP*GP*TP*GP*TP*GP*AP*G )-3'), DNA (5'-D(*CP*TP*CP*AP*CP*AP*CP*GP*TP*GP*GP*GP*AP*CP*TP*AP*G )-3'), PROTEIN (PHOSPHATE SYSTEM POSITIVE REGULATORY PROTEIN PHO4) | | Authors: | Shimizu, T, Toumoto, A, Ihara, K, Shimizu, M, Kyogoku, Y, Ogawa, N, Oshima, Y, Hakoshima, T. | | Deposit date: | 1997-11-27 | | Release date: | 1998-03-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of PHO4 bHLH domain-DNA complex: flanking base recognition.
EMBO J., 16, 1997
|
|
1ISN
 
 | | Crystal structure of merlin FERM domain | | Descriptor: | merlin | | Authors: | Shimizu, T, Seto, A, Maita, N, Hamada, K, Tsukita, S, Tsukita, S, Hakoshima, T. | | Deposit date: | 2001-12-13 | | Release date: | 2002-04-03 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for neurofibromatosis type 2. Crystal structure of the merlin FERM domain.
J.Biol.Chem., 277, 2002
|
|
1RLS
 
 | | CRYSTAL STRUCTURE OF RNASE T1 COMPLEXED WITH THE PRODUCT NUCLEOTIDE 3'-GMP. STRUCTURAL EVIDENCE FOR DIRECT INTERACTION OF HISTIDINE 40 AND GLUTAMIC ACID 58 WITH THE 2'-HYDROXYL GROUP OF RIBOSE | | Descriptor: | CALCIUM ION, GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Gohda, K, Oka, K.-I, Tomita, K.-I, Hakoshima, T. | | Deposit date: | 1994-03-29 | | Release date: | 1994-12-20 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of RNase T1 complexed with the product nucleotide 3'-GMP. Structural evidence for direct interaction of histidine 40 and glutamic acid 58 with the 2'-hydroxyl group of the ribose.
J.Biol.Chem., 269, 1994
|
|
5B3H
 
 | | The crystal structure of the JACKDAW/IDD10 bound to the heterodimeric SHR-SCR complex | | Descriptor: | Protein SCARECROW, Protein SHORT-ROOT, ZINC ION, ... | | Authors: | Hirano, Y, Suyama, T, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
5B5W
 
 | | Crystal structure of MOB1-LATS1 NTR domain complex | | Descriptor: | MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.957 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B6B
 
 | | Complex of LATS1 and phosphomimetic MOB1b | | Descriptor: | CHLORIDE ION, MOB kinase activator 1B, Serine/threonine-protein kinase LATS1, ... | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.536 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B5V
 
 | | Structure of full-length MOB1b | | Descriptor: | CHLORIDE ION, MOB kinase activator 1B, ZINC ION | | Authors: | KIM, S.-Y, Tachioka, Y, Mori, T, Hakoshima, T. | | Deposit date: | 2016-05-24 | | Release date: | 2016-07-06 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.193 Å) | | Cite: | Structural basis for autoinhibition and its relief of MOB1 in the Hippo pathway
Sci Rep, 6, 2016
|
|
5B3G
 
 | | The crystal structure of the heterodimer of SHORT-ROOT and SCARECROW GRAS domains | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Hirano, Y, Nakagawa, M, Hakoshima, T. | | Deposit date: | 2016-02-29 | | Release date: | 2017-03-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the SHR-SCR heterodimer bound to the BIRD/IDD transcriptional factor JKD
Nat Plants, 3, 2017
|
|
293D
 
 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE-2: THE CRYSTAL STRUCTURE OF THE D(CG)3 AND SPERMIDINE COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, SODIUM ION, ... | | Authors: | Ohishi, H, Nakanishi, I, Inubushi, K, Van Der Marel, G.A, Van Boom, J.H, Rich, A, Wang, A.H.-J, Hakoshima, T, Tomita, K. | | Deposit date: | 1996-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine-2. The crystal structure of the d(CG)3 and spermidine complex.
FEBS Lett., 391, 1996
|
|
292D
 
 | | INTERACTION BETWEEN THE LEFT-HANDED Z-DNA AND POLYAMINE:THE CRYSTAL STRUCTURE OF THE D(CG)3 AND N-(2-AMINOETHYL)-1,4-DIAMINOBUTANE COMPLEX | | Descriptor: | 1-(AMINOETHYL)AMINO-4-AMINOBUTANE, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION, ... | | Authors: | Ohishi, H, Kunisawa, S, Van Der Marel, G, Van Boom, J.H, Rich, A, Wang, A.H.-J, Tomita, K, Hakoshima, T. | | Deposit date: | 1991-10-09 | | Release date: | 1996-12-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Interaction between the left-handed Z-DNA and polyamine. The crystal structure of the d(CG)3 and N-(2-aminoethyl)-1,4-diamino-butane complex.
FEBS Lett., 284, 1991
|
|
5YIZ
 
 | |
5YJ0
 
 | |
5YJ1
 
 | | Mouse Cereblon thalidomide binding domain complexed with R-form thalidomide | | Descriptor: | 2-[(3~{R})-2,6-bis(oxidanylidene)piperidin-3-yl]isoindole-1,3-dione, Protein cereblon, SULFATE ION, ... | | Authors: | Mori, T, Hakoshima, T. | | Deposit date: | 2017-10-06 | | Release date: | 2018-02-07 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of thalidomide enantiomer binding to cereblon
Sci Rep, 8, 2018
|
|
5XAV
 
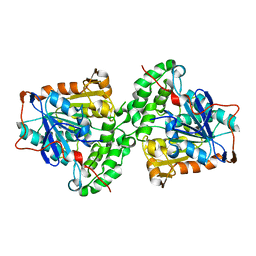 | | Structure of PhaC from Chromobacterium sp. USM2 | | Descriptor: | Intracellular polyhydroxyalkanoate synthase | | Authors: | Chek, M.F, Kim, S.Y, Mori, T, Arsad, H, Samian, M.R, Sudesh, K, Hakoshima, T. | | Deposit date: | 2017-03-15 | | Release date: | 2017-07-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.479 Å) | | Cite: | Structure of polyhydroxyalkanoate (PHA) synthase PhaC from Chromobacterium sp. USM2, producing biodegradable plastics
Sci Rep, 7, 2017
|
|
7DCH
 
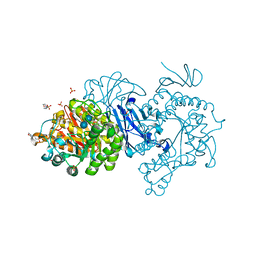 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with acarbose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Alpha-glycosidase, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9C
 
 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-13 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7D9B
 
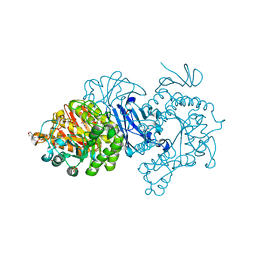 | | Crystal structure of alpha-glucosidase | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-12 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DCG
 
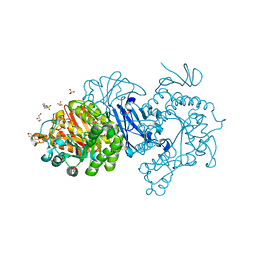 | | Alpha-glucosidase from Weissella cibaria BBK-1 bound with maltotriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Alpha-glycosidase, CALCIUM ION, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2020-10-26 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EHI
 
 | | Crystal structure of covalent maltosyl-alpha-glucosidase intermediate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7EHH
 
 | | Crystal structure of alpha-glucosidase from Weissella cibaria BKK1 in complex with maltose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Krusong, K, Wangpaiboon, K, Kim, S, Mori, T, Hakoshima, T. | | Deposit date: | 2021-03-29 | | Release date: | 2021-08-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A GH13 alpha-glucosidase from Weissella cibaria uncommonly acts on short-chain maltooligosaccharides.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
6KYW
 
 | | S8-mSRK-S8-SP11 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Receptor protein kinase SRK8, S locus protein 11 | | Authors: | Murase, K, Hakoshima, T, Mori, T. | | Deposit date: | 2019-09-20 | | Release date: | 2020-09-16 | | Last modified: | 2020-10-21 | | Method: | X-RAY DIFFRACTION (2.60112739 Å) | | Cite: | Mechanism of self/nonself-discrimination in Brassica self-incompatibility.
Nat Commun, 11, 2020
|
|
