3W35
 
 | | Crystal structure of apo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1 | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
3W36
 
 | | Crystal structure of holo-type bacterial Vanadium-dependent chloroperoxidase | | Descriptor: | NapH1, VANADATE ION | | Authors: | Liscombe, D.K, Miyanaga, A, Fielding, E, Bernhardt, P, Li, A, Winter, J.M, Gilson, M.K, Noel, J.P, Moore, B.S. | | Deposit date: | 2012-12-11 | | Release date: | 2013-12-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural Basis of Stereospecific Vanadium-Dependent Haloperoxidase Family Enzymes in Napyradiomycin Biosynthesis.
Biochemistry, 2022
|
|
6XRL
 
 | | Crystal structure of human PI3K-gamma in complex with inhibitor IPI-549 | | Descriptor: | 2-amino-N-[(1S)-1-{8-[(1-methyl-1H-pyrazol-4-yl)ethynyl]-1-oxo-2-phenyl-1,2-dihydroisoquinolin-3-yl}ethyl]pyrazolo[1,5-a]pyrimidine-3-carboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
6XRM
 
 | | Crystal structure of human PI3K-gamma in complex with Compound 4 | | Descriptor: | 5-[2-amino-3-(1-methyl-1H-pyrazol-4-yl)pyrazolo[1,5-a]pyrimidin-5-yl]-2-[(1S)-1-cyclopropylethyl]-7-(trifluoromethyl)-2,3-dihydro-1H-isoindol-1-one, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Walker, N.P, Jeffrey, J.L. | | Deposit date: | 2020-07-13 | | Release date: | 2020-09-16 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Discovery of Potent and Selective PI3K gamma Inhibitors.
J.Med.Chem., 63, 2020
|
|
7SUC
 
 | |
5C8A
 
 | |
5C8E
 
 | |
5C8D
 
 | |
5C8F
 
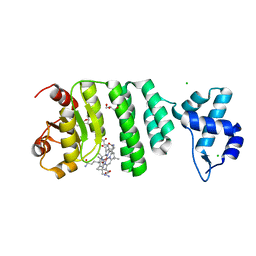 | |
8I86
 
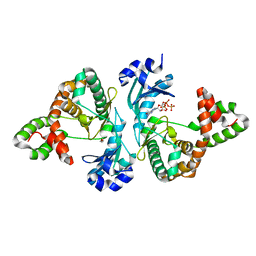 | | Crystal structure of Cph001-D189N in complex with GTP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-TRIPHOSPHATE, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I84
 
 | | Crystal structure of Cph001-D189N in complex with CMN IIB | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, KBE-DPP-UAL-MYN-DPP-ALA, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I85
 
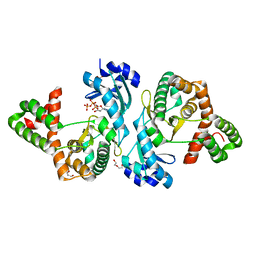 | | Crystal structure of Cph001-D189N in complex with ATP | | Descriptor: | ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I89
 
 | | Crystal structure of Cph001-D189N in complex with VIO | | Descriptor: | DI(HYDROXYETHYL)ETHER, KBE-DPP-SER-SER-UAL-5OH, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-03 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I8G
 
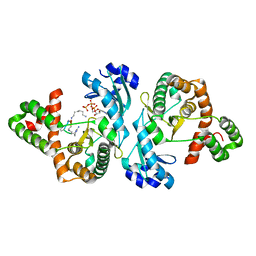 | | Crystal structure of Cph001-D189N in complex with CMN IIA and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, KBE-DPP-UAL-MYN-DPP-SER, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I8H
 
 | | Crystal structure of Cph001-D189N in complex with VIO and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DI(HYDROXYETHYL)ETHER, KBE-DPP-SER-SER-UAL-5OH, ... | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J, Hsiao, P.Y. | | Deposit date: | 2023-02-04 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I80
 
 | | Crystal structure of Cph001-D189N | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, Viomycin kinase | | Authors: | Chang, C.Y, Toh, S.I, Elaine K, J. | | Deposit date: | 2023-02-02 | | Release date: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery and characterization of genes conferring natural resistance to the antituberculosis antibiotic capreomycin.
Commun Biol, 6, 2023
|
|
8I82
 
 | |
6JZ0
 
 | | Crystal structure of EGFR kinase domain in complex with compound 78 | | Descriptor: | E-4-(dimethylamino)-N-[3-[4-[[(1S)-2-oxidanyl-1-phenyl-ethyl]amino]-6-phenyl-furo[2,3-d]pyrimidin-5-yl]phenyl]but-2-enamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S, Wu, S.Y. | | Deposit date: | 2019-04-30 | | Release date: | 2019-12-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Discovery of a Furanopyrimidine-Based Epidermal Growth Factor Receptor Inhibitor (DBPR112) as a Clinical Candidate for the Treatment of Non-Small Cell Lung Cancer.
J.Med.Chem., 62, 2019
|
|
5Z19
 
 | |
5Z1A
 
 | |
5Z18
 
 | |
5Z1B
 
 | |
7EI3
 
 | |
7EI4
 
 | | Crystal structure of MasL in complex with a novel covalent inhibitor, collimonin C | | Descriptor: | (6S,7R,9E)-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-03-30 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
7FEA
 
 | | PY14 in complex with Col-D | | Descriptor: | (6~{R},7~{R},9~{E})-6,7-bis(oxidanyl)hexadeca-9,15-dien-11,13-diynoic acid, Acetyl-CoA C-acyltransferase | | Authors: | Lin, C.C, Ko, T.P, Huang, K.F, Yang, Y.L. | | Deposit date: | 2021-07-19 | | Release date: | 2022-07-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Integrated omics approach to unveil antifungal bacterial polyynes as acetyl-CoA acetyltransferase inhibitors.
Commun Biol, 5, 2022
|
|
