1VBZ
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in Ba2+ solution | | Descriptor: | BARIUM ION, Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VBY
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, and Mn2+ bound | | Descriptor: | Hepatitis Delta virus ribozyme, MANGANESE (II) ION, SODIUM ION, ... | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VC6
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Product with C75U Mutaion, cleaved in Imidazole and Mg2+ solutions | | Descriptor: | Hepatitis Delta virus ribozyme, MAGNESIUM ION, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-04 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
1VBX
 
 | | Crystal Structure of the Hepatitis Delta Virus Gemonic Ribozyme Precursor, with C75U mutaion, in EDTA solution | | Descriptor: | Hepatitis Delta virus ribozyme, U1 small nuclear ribonucleoprotein A | | Authors: | Ke, A, Zhou, K, Ding, F, Cate, J.H.D, Doudna, J.A. | | Deposit date: | 2004-03-03 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A Conformational Switch controls hepatitis delta virus ribozyme catalysis
NATURE, 429, 2004
|
|
4V9C
 
 | | Allosteric control of the ribosome by small-molecule antibiotics | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Cate, J.H.D, Pulk, A, Blanchard, S.C, Wang, L, Feldman, M.B, Wasserman, M.R, Altman, R. | | Deposit date: | 2012-07-25 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Allosteric control of the ribosome by small-molecule antibiotics.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6XA1
 
 | |
7K00
 
 | | Structure of the Bacterial Ribosome at 2 Angstrom Resolution | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Watson, Z.L, Ward, F.R, Meheust, R, Ad, O, Schepartz, A, Banfield, J.F, Cate, J.H.D. | | Deposit date: | 2020-09-02 | | Release date: | 2020-09-23 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structure of the bacterial ribosome at 2 angstrom resolution.
Elife, 9, 2020
|
|
6I7V
 
 | | Ribosomal protein paralogs bL31 and bL36 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Pulk, A, Cate, J.H.D, Remme, J, Lilleorg, S, Reier, K, Peil, L, Liiv, A, Tammsalu, T. | | Deposit date: | 2018-11-19 | | Release date: | 2018-12-05 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Bacterial ribosome heterogeneity: Changes in ribosomal protein composition during transition into stationary growth phase.
Biochimie, 156, 2018
|
|
4YBB
 
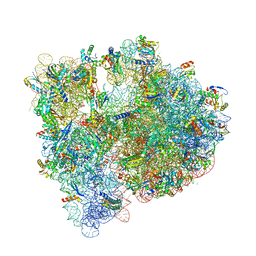 | | High-resolution structure of the Escherichia coli ribosome | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Noeske, J, Wasserman, M.R, Terry, D.S, Altman, R.B, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2015-02-18 | | Release date: | 2015-03-18 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | High-resolution structure of the Escherichia coli ribosome.
Nat.Struct.Mol.Biol., 22, 2015
|
|
9DUK
 
 | |
9DUL
 
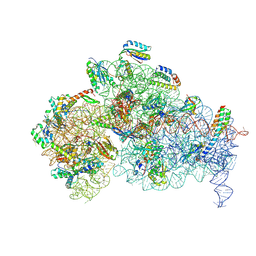 | |
4V6C
 
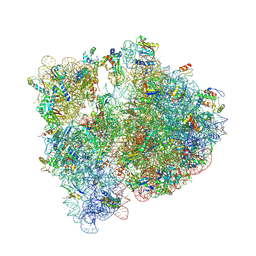 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V7V
 
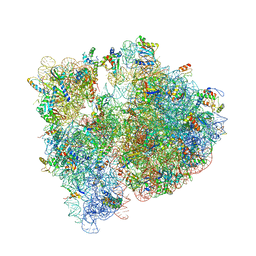 | | Crystal structure of the E. coli ribosome bound to clindamycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-16 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2891 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
5K4B
 
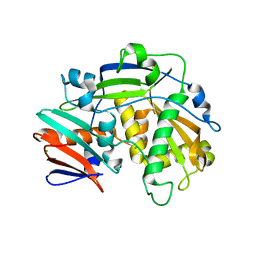 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 1 | | Descriptor: | CHLORIDE ION, Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.399 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4D
 
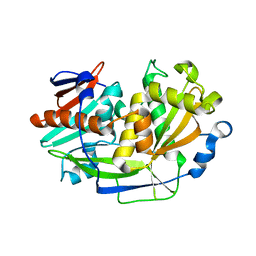 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4C
 
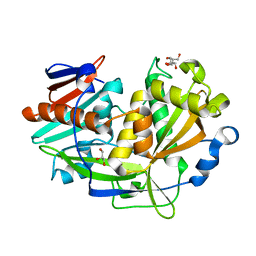 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 2 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D, GLYCEROL | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.698 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
3IY9
 
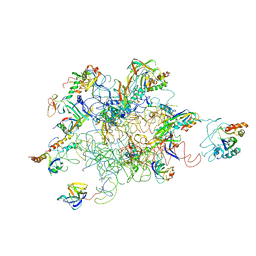 | | Leishmania Tarentolae Mitochondrial Large Ribosomal Subunit Model | | Descriptor: | 39S ribosomal protein L11, mitochondrial, 39S ribosomal protein L16, ... | | Authors: | Sharma, M.R, Booth, T.M, Simpson, L, Maslov, D.A, Agrawal, R.K. | | Deposit date: | 2009-04-20 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (14.1 Å) | | Cite: | Structure of a mitochondrial ribosome with minimal RNA
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
4V9D
 
 | | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Wang, L, Feldman, M.B, Pulk, A, Chen, V.B, Kapral, G.J, Noeske, J, Richardson, J.S, Blanchard, S.C, Cate, J.H.D. | | Deposit date: | 2012-07-31 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structures of the bacterial ribosome in classical and hybrid states of tRNA binding.
Science, 332, 2011
|
|
4V54
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-16 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V6E
 
 | | Crystal structure of the E. coli 70S ribosome in an intermediate state of ratcheting | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Zhang, W, Dunkle, J.A, Cate, J.H.D. | | Deposit date: | 2009-06-28 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.712 Å) | | Cite: | Structures of the ribosome in intermediate States of ratcheting.
Science, 325, 2009
|
|
4V4Q
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli at 3.5 A resolution. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Schuwirth, B.S, Borovinskaya, M.A, Hau, C.W, Zhang, W, Vila-Sanjurjo, A, Holton, J.M, Cate, J.H.D. | | Deposit date: | 2005-08-30 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.46 Å) | | Cite: | Structures of the bacterial ribosome at 3.5 A resolution.
Science, 310, 2005
|
|
4V9O
 
 | |
4V5Y
 
 | | Crystal structure of the bacterial ribosome from Escherichia coli in complex with paromomycin and ribosome recycling factor (RRF). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Borovinskaya, M.A, Pai, R.D, Zhang, W, Schuwirth, B.-S, Holton, J.M, Hirokawa, G, Kaji, H, Kaji, A, Cate, J.H.D. | | Deposit date: | 2007-06-19 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (4.45 Å) | | Cite: | Structural basis for aminoglycoside inhibition of bacterial ribosome recycling.
Nat.Struct.Mol.Biol., 14, 2007
|
|
4V7S
 
 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4V7T
 
 | | Crystal structure of the E. coli ribosome bound to chloramphenicol. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-14 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.1942 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
