7P3Q
 
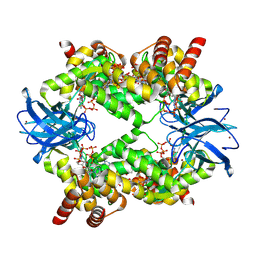 | | Streptomyces coelicolor dATP/ATP-loaded NrdR octamer | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ... | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-08 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
7P3F
 
 | |
7P37
 
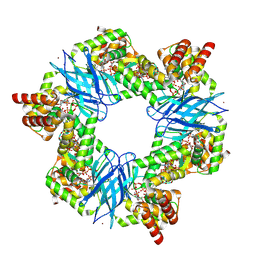 | | Streptomyces coelicolor ATP-loaded NrdR | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Transcriptional repressor NrdR, ZINC ION | | Authors: | Martinez-Carranza, M, Stenmark, P. | | Deposit date: | 2021-07-07 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases.
Nat Commun, 13, 2022
|
|
6TQL
 
 | | Cryo-EM of elastase-treated human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
6TQK
 
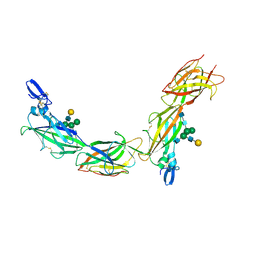 | | Cryo-EM of native human uromodulin (UMOD)/Tamm-Horsfall protein (THP) filament. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Uromodulin, ... | | Authors: | Stsiapanava, A, Xu, C, Carroni, M, Wu, B, Jovine, L. | | Deposit date: | 2019-12-16 | | Release date: | 2020-11-04 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Embo J., 39, 2020
|
|
4X0J
 
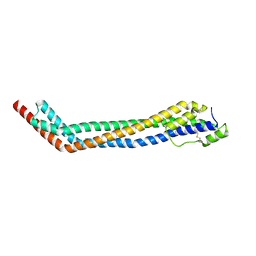 | | Trypanosoma brucei haptoglobin-haemoglobin receptor | | Descriptor: | Haptoglobin-hemoglobin receptor | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
7QFQ
 
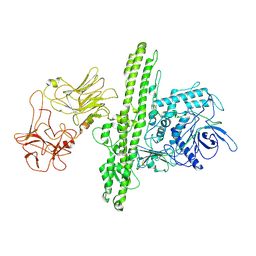 | | Cryo-EM structure of Botulinum neurotoxin serotype B | | Descriptor: | Botulinum neurotoxin type B | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
7QFP
 
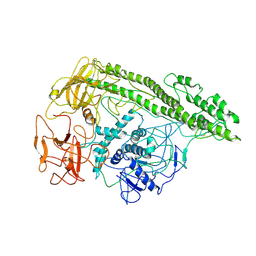 | | Cryo-EM structure of Botulinum neurotoxin serotype E | | Descriptor: | Botulinum neurotoxin | | Authors: | Kosenina, S, Martinez-Carranza, M, Davies, J.R, Masuyer, G, Stenmark, P. | | Deposit date: | 2021-12-06 | | Release date: | 2022-01-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural Analysis of Botulinum Neurotoxins Type B and E by Cryo-EM.
Toxins, 14, 2021
|
|
4X0L
 
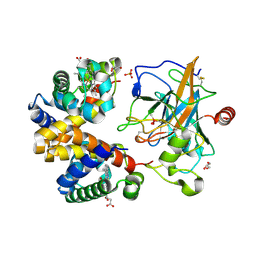 | | Human haptoglobin-haemoglobin complex | | Descriptor: | CACODYLATE ION, GLYCEROL, Haptoglobin, ... | | Authors: | Lane-Serff, H, MacGregor, P, Lowe, E.D, Carrington, M, Higgins, M.K. | | Deposit date: | 2014-11-21 | | Release date: | 2014-12-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for ligand and innate immunity factor uptake by the trypanosome haptoglobin-haemoglobin receptor.
Elife, 3, 2014
|
|
7NTM
 
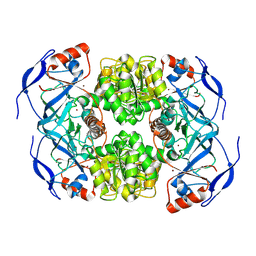 | |
6HMS
 
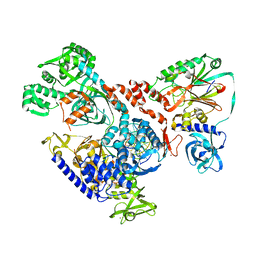 | | Cryo-EM map of DNA polymerase D from Pyrococcus abyssi in complex with DNA | | Descriptor: | DNA (5'-D(*GP*AP*GP*AP*CP*GP*GP*GP*CP*CP*GP*CP*GP*TP*C)-3'), DNA (5'-D(P*TP*GP*AP*CP*GP*CP*GP*GP*CP*CP*CP*GP*TP*CP*TP*C)-3'), DNA polymerase II large subunit,DNA polymerase II large subunit, ... | | Authors: | Raia, P, Carroni, M, Sauguet, L. | | Deposit date: | 2018-09-12 | | Release date: | 2019-01-30 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.1 Å) | | Cite: | Structure of the DP1-DP2 PolD complex bound with DNA and its implications for the evolutionary history of DNA and RNA polymerases.
PLoS Biol., 17, 2019
|
|
8C6C
 
 | | Light SFX structure of D.m(6-4)photolyase at 300ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6H
 
 | | Light SFX structure of D.m(6-4)photolyase at 2ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C1U
 
 | | SFX structure of D.m(6-4)photolyase | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2022-12-21 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C69
 
 | | Light SFX structure of D.m(6-4)photolyase at 100 microsecond time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6A
 
 | | Light SFX structure of D.m(6-4)photolyase at 1ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6B
 
 | | Light SFX structure of D.m(6-4)photolyase at 20ps time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
8C6F
 
 | | Light SFX structure of D.m(6-4)photolyase at 400fs time delay | | Descriptor: | Cryptochrome-1, FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL | | Authors: | Cellini, A, Kumar, M, Nimmrich, A, Mutisya, J, Furrer, A, Beale, E.V, Carrillo, M, Malla, T.N, Maj, P, Dworkowskic, F, Cirelli, C, Ozerovi, D, Bacellar, C, Strandfuss, J, Weinert, T, Ihalainen, J.A, Yuan Wahlgren, W, Westenhoff, S. | | Deposit date: | 2023-01-11 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Directed ultrafast conformational changes accompany electron transfer in a photolyase as resolved by serial crystallography.
Nat.Chem., 16, 2024
|
|
6FLS
 
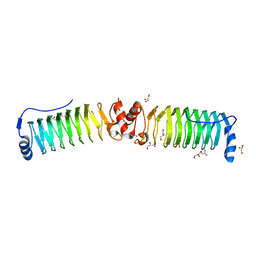 | |
5FTU
 
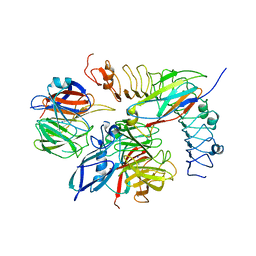 | | Tetrameric complex of Latrophilin 3, Unc5D and FLRT2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADHESION G PROTEIN-COUPLED RECEPTOR L3, CALCIUM ION, ... | | Authors: | Jackson, V.A, Mehmood, S, Chavent, M, Roversi, P, Carrasquero, M, del Toro, D, Seyit-Bremer, G, Ranaivoson, F.M, Comoletti, D, Sansom, M.S.P, Robinson, C.V, Klein, R, Seiradake, E. | | Deposit date: | 2016-01-15 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.01 Å) | | Cite: | Super-Complexes of Adhesion Gpcrs and Neural Guidance Receptors
Nat.Commun., 7, 2016
|
|
5AFB
 
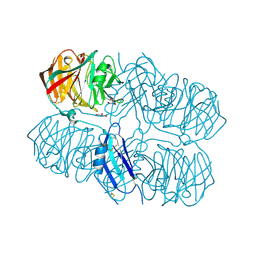 | | Crystal structure of the Latrophilin3 Lectin and Olfactomedin Domains | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, GLYCEROL, ... | | Authors: | Jackson, V.A, del Toro, D, Carrasquero, M, Roversi, P, Harlos, K, Klein, R, Seiradake, E. | | Deposit date: | 2015-01-21 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structural Basis of Latrophilin-Flrt Interaction.
Structure, 23, 2015
|
|
5OFO
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
5OG1
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state | | Descriptor: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | Authors: | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | Deposit date: | 2017-07-11 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
6ZGK
 
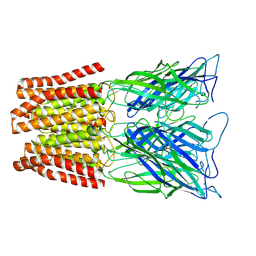 | | GLIC pentameric ligand-gated ion channel, pH 3 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
6ZGD
 
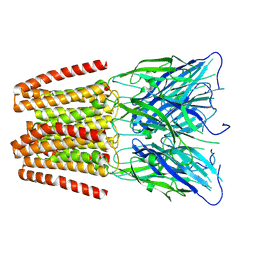 | | GLIC pentameric ligand-gated ion channel, pH 7 | | Descriptor: | Proton-gated ion channel | | Authors: | Rovsnik, U, Zhuang, Y, Forsberg, B.O, Carroni, M, Yvonnesdotter, L, Howard, R.J, Lindahl, E. | | Deposit date: | 2020-06-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Dynamic closed states of a ligand-gated ion channel captured by cryo-EM and simulations.
Life Sci Alliance, 4, 2021
|
|
