7AU7
 
 | | Crystal structure of Nod Factor Perception ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Serine/threonine receptor-like kinase NFP, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-11-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.547 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4UZ3
 
 | | Crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from T. thermophilus bound to N-acetyl-chitohexaose | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, ... | | Authors: | Wong, J.E.M.M, Blaise, M. | | Deposit date: | 2014-09-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An Intermolecular Binding Mechanism Involving Multiple Lysm Domains Mediates Carbohydrate Recognition by an Endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8QNN
 
 | | Crystal structure of a Class A beta-lactamase from Nocardia cyriacigeorgica | | Descriptor: | Beta-lactamase, CITRATE ANION | | Authors: | Feuillard, J, Couston, J, Benito, Y, Hodille, E, Dumitrescu, O, Blaise, M. | | Deposit date: | 2023-09-27 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Biochemical and structural characterization of a class A beta-lactamase from Nocardia cyriacigeorgica.
Acta Crystallogr.,Sect.F, 80, 2024
|
|
8QKK
 
 | | Cryo-EM structure of MmpL3 from Mycobacterium smegmatis reconstituted into peptidiscs | | Descriptor: | Trehalose monomycolate exporter MmpL3 | | Authors: | Couston, J, Guo, Z, Wang, K, Gourdon, P.E, Blaise, M. | | Deposit date: | 2023-09-15 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Cryo-EM structure of the trehalose monomycolate transporter, MmpL3, reconstituted into peptidiscs.
Curr Res Struct Biol, 6, 2023
|
|
8CDA
 
 | | Crystal structure of MAB_4123 from Mycobacterium abscessus | | Descriptor: | GLYCEROL, PHOSPHATE ION, Probable monooxygenase | | Authors: | Ung, K.L, Poussineau, C, Couston, J, Alsarraf, H.M.A.B, Blaise, M. | | Deposit date: | 2023-01-30 | | Release date: | 2023-05-10 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of MAB_4123, a putative flavin-dependent monooxygenase from Mycobacterium abscessus.
Acta Crystallogr.,Sect.F, 79, 2023
|
|
7BAX
 
 | | Crystal structure of LYS11 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase | | Authors: | Laursen, M, Cheng, J, Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-12-16 | | Release date: | 2021-11-10 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Kinetic proofreading of lipochitooligosaccharides determines signal activation of symbiotic plant receptors.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5LS2
 
 | | Receptor mediated chitin perception in legumes is functionally seperable from Nod factor perception | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LysM type receptor kinase, SULFATE ION | | Authors: | Bozsoki, Z, Cheng, J, Feng, F, Gysel, K, Andersen, K.R, Oldroyd, G, Blaise, M, Radutoiu, S, Stougaard, J. | | Deposit date: | 2016-08-22 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Receptor-mediated chitin perception in legume roots is functionally separable from Nod factor perception.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6EZE
 
 | | The open conformation of E.coli Elongation Factor Tu in complex with GDPNP. | | Descriptor: | DI(HYDROXYETHYL)ETHER, Elongation factor Tu 2, GLYCEROL, ... | | Authors: | Johansen, J.S, Blaise, M, Thirup, S.S. | | Deposit date: | 2017-11-15 | | Release date: | 2018-08-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | E. coli elongation factor Tu bound to a GTP analogue displays an open conformation equivalent to the GDP-bound form.
Nucleic Acids Res., 46, 2018
|
|
1N9W
 
 | | Crystal structure of the non-discriminating and archaeal-type aspartyl-tRNA synthetase from Thermus thermophilus | | Descriptor: | aspartyl-tRNA synthetase 2 | | Authors: | Charron, C, Roy, H, Blaise, M, Giege, R, Kern, D. | | Deposit date: | 2002-11-26 | | Release date: | 2003-04-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Non-discriminating and discriminating aspartyl-tRNA synthetases differ in
the anticodon-binding domain
EMBO J., 22, 2003
|
|
6XWE
 
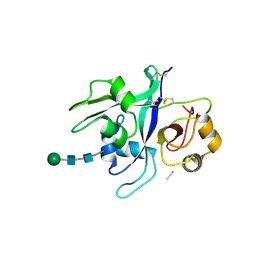 | | Crystal structure of LYK3 ectodomain | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ACETONITRILE, LysM domain receptor-like kinase 3, ... | | Authors: | Gysel, K, Blaise, M, Andersen, K.R. | | Deposit date: | 2020-01-23 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Ligand-recognizing motifs in plant LysM receptors are major determinants of specificity.
Science, 369, 2020
|
|
4UZ2
 
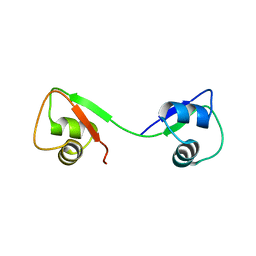 | |
4XCM
 
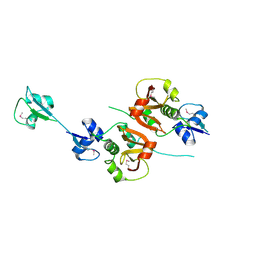 | | Crystal structure of the putative NlpC/P60 D,L endopeptidase from T. thermophilus | | Descriptor: | Cell wall-binding endopeptidase-related protein | | Authors: | Wong, J, Midtgaard, S, Gysel, K, Thygesen, M.B, Sorensen, K.K, Jensen, K.J, Stougaard, J, Thirup, S, Blaise, M. | | Deposit date: | 2014-12-18 | | Release date: | 2015-01-14 | | Last modified: | 2018-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | An intermolecular binding mechanism involving multiple LysM domains mediates carbohydrate recognition by an endopeptidase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5OVK
 
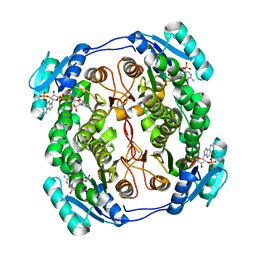 | | Crystal structure MabA bound to NADPH from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OVL
 
 | | crystal structure of MabA bound to NADP+ from M. smegmatis | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OCJ
 
 | | Crystal structure of Ag85C bound to cyclophostin 8beta inhibitor | | Descriptor: | DIMETHYL SULFOXIDE, Diacylglycerol acyltransferase/mycolyltransferase Ag85C, methoxy-[(3~{R})-3-[(2~{R})-1-methoxy-1,3-bis(oxidanylidene)butan-2-yl]pentadecyl]phosphinic acid | | Authors: | Viljoen, A, Richard, M, Nguyen, P.C, Spilling, C.D, Canaan, S, Cavalier, J.F, Blaise, M, Kremer, L. | | Deposit date: | 2017-07-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Cyclipostins and cyclophostin analogs inhibit the antigen 85C from
J. Biol. Chem., 293, 2018
|
|
5OVJ
 
 | | Structure of the apo form of Mycobacterium smegmatis MabA | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase FabG, SULFATE ION | | Authors: | Kussau, T, Van Wyk, N, Viljoen, A, Olieric, V, Flipo, M, Kremer, L, Blaise, M. | | Deposit date: | 2017-08-29 | | Release date: | 2018-02-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural rearrangements occurring upon cofactor binding in the Mycobacterium smegmatis beta-ketoacyl-acyl carrier protein reductase MabA.
Acta Crystallogr D Struct Biol, 74, 2018
|
|
5OVY
 
 | | Crystal structure of MAB_4384 tetR | | Descriptor: | Putative transcriptional regulator, TetR family | | Authors: | Richard, M, Gutierrez, A.V, Viljoen, A, Ghigo, E, Blaise, M, Kremer, L. | | Deposit date: | 2017-08-30 | | Release date: | 2018-03-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic and Structural Insights Into the Unique TetR-Dependent Regulation of a Drug Efflux Pump inMycobacterium abscessus.
Front Microbiol, 9, 2018
|
|
6QUB
 
 | | Truncated beta-galactosidase III from Bifidobacterium bifidum in complex with galactose | | Descriptor: | Beta-galactosidase, CALCIUM ION, beta-D-galactopyranose | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUC
 
 | | Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | Beta-galactosidase, CALCIUM ION, IMIDAZOLE | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
6QUD
 
 | | 2-deoxy-galactose reaction intermediate of a Truncated beta-galactosidase III from Bifidobacterium bifidum | | Descriptor: | 2-deoxy-alpha-D-galactopyranose, Beta-galactosidase, CALCIUM ION | | Authors: | Thirup, S.S, Nielsen, J.A, Andersen, J.L, Alsarraf, H, Blaise, M. | | Deposit date: | 2019-02-27 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Truncated beta-galactosidase III from Bifidobacterium bifidum
To Be Published
|
|
5T2U
 
 | | short chain dehydrogenase/reductase family protein | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, short chain dehydrogenase/reductase family protein | | Authors: | Mickael, B, Van Wyk, N, Baneres-Roquet, F, Yann, G, Laurent, K. | | Deposit date: | 2016-08-24 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Binding of NADP(+) triggers an open-to-closed transition in a mycobacterial FabG beta-ketoacyl-ACP reductase.
Biochem. J., 474, 2017
|
|
