3JS3
 
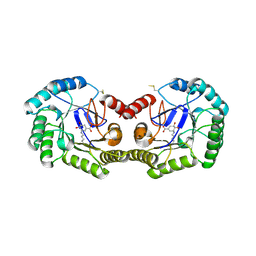 | | Crystal structure of type I 3-dehydroquinate dehydratase (aroD) from Clostridium difficile with covalent reaction intermediate | | Descriptor: | 3-AMINO-4,5-DIHYDROXY-CYCLOHEX-1-ENECARBOXYLATE, 3-dehydroquinate dehydratase | | Authors: | Minasov, G, Light, S.H, Shuvalova, L, Dubrovska, I, Winsor, J, Peterson, S.N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-09 | | Release date: | 2009-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Insights into the mechanism of type I dehydroquinate dehydratases from structures of reaction intermediates.
J.Biol.Chem., 286, 2011
|
|
5JPH
 
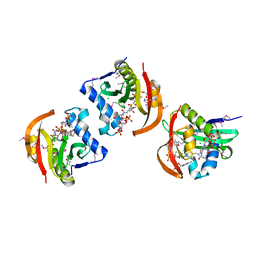 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus in complex with CoA | | Descriptor: | Acetyltransferase SACOL1063, CHLORIDE ION, COENZYME A | | Authors: | Majorek, K.A, Osinski, T, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-03 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
5JOQ
 
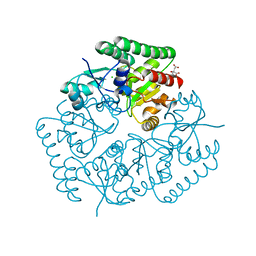 | | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e | | Descriptor: | CHLORIDE ION, CITRIC ACID, Lmo2184 protein | | Authors: | Brunzelle, J.S, Wawrzak, Z, Kudritska, M, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-02 | | Release date: | 2016-07-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal Structure of an ABC Transporter Substrate-Binding Protein from Listeria monocytogenes EGD-e
To Be Published
|
|
5JQC
 
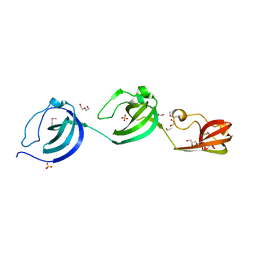 | | Crystal structure putative autolysin from Listeria monocytogenes | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Lmo1076 protein, ... | | Authors: | Chang, C, Zhou, M, Shatsman, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-05-18 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Crystal structure putative autolysin from Listeria monocytogenes
To Be Published
|
|
5JTH
 
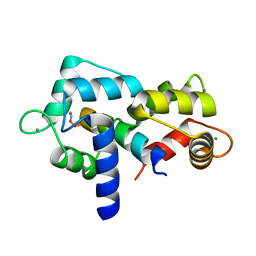 | | Crystal structure of E67A calmodulin - CaM:RM20 analog complex | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase, ... | | Authors: | Grum-Tokars, V.L, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2016-05-09 | | Release date: | 2017-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal structure of E67A calmodulin - CaM:RM20 analog complex
To Be Published
|
|
5JQA
 
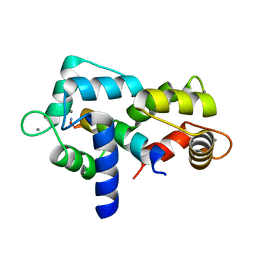 | | CaM:RM20 complex | | Descriptor: | CALCIUM ION, Calmodulin, Myosin light chain kinase, ... | | Authors: | Tokars, V.L, Minasov, G, Anderson, W.F, Watterson, D.M. | | Deposit date: | 2016-05-04 | | Release date: | 2017-10-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CaM:RM20 complex
To be Published
|
|
5K9X
 
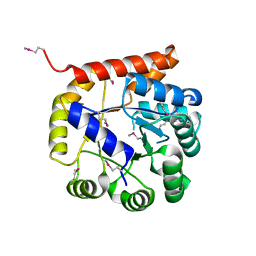 | | Crystal structure of Tryptophan synthase alpha chain from Legionella pneumophila subsp. pneumophila | | Descriptor: | Tryptophan synthase alpha chain | | Authors: | Chang, C, Hatzos-Skintges, C, Endres, M, ANDERSON, W.F, JOACHIMIAK, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-06-01 | | Release date: | 2016-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.016 Å) | | Cite: | Crystal structure of Tryptophan synthase alpha chain from Legionella pneumophila subsp. pneumophila
To Be Published
|
|
5JQ4
 
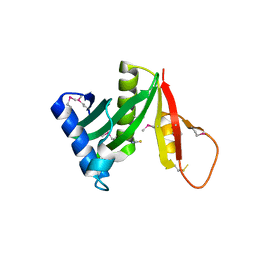 | | Structure of a GNAT acetyltransferase SACOL1063 from Staphylococcus aureus | | Descriptor: | 1,2-ETHANEDIOL, Acetyltransferase SACOL1063, CHLORIDE ION, ... | | Authors: | Majorek, K.A, Anderson, W.F, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-05-04 | | Release date: | 2016-06-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Insight into the 3D structure and substrate specificity of previously uncharacterized GNAT superfamily acetyltransferases from pathogenic bacteria.
Biochim.Biophys.Acta, 1865, 2016
|
|
5DIB
 
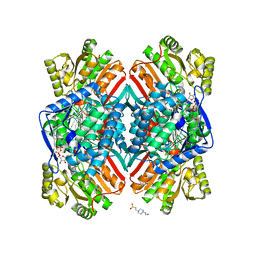 | | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | 2.25 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) Y450L point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
3SB1
 
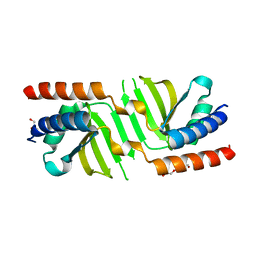 | | Hydrogenase expression protein HupH from Thiobacillus denitrificans ATCC 25259 | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, hydrogenase expression protein | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Ruan, J, Leo, R.D, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-06-03 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Hydrogenase expression protein HupH from Thiobacillus denitrificans ATCC 25259
To be Published
|
|
3S7Z
 
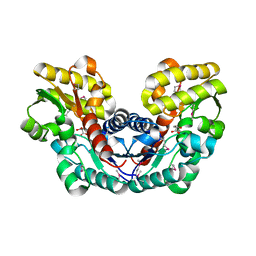 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate | | Descriptor: | MAGNESIUM ION, Putative aspartate racemase, SUCCINIC ACID, ... | | Authors: | Maltseva, N, Zhang, R, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-10-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium Complexed with Succinate.
To be Published
|
|
3S81
 
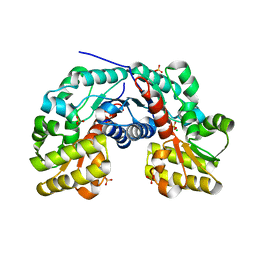 | | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium | | Descriptor: | CHLORIDE ION, Putative aspartate racemase, SULFATE ION | | Authors: | Maltseva, N, Kim, Y, Kwon, K, Zhang, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.796 Å) | | Cite: | Crystal Structure of Putative Aspartate Racemase from Salmonella Typhimurium
To be Published
|
|
3S52
 
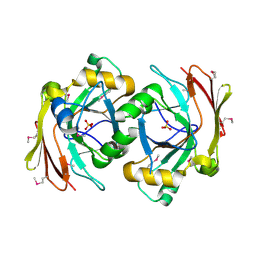 | | Crystal structure of a putative fumarylacetoacetate hydrolase family protein from Yersinia pestis CO92 | | Descriptor: | CHLORIDE ION, Putative fumarylacetoacetate hydrolase family protein, SULFATE ION | | Authors: | Nocek, B, Zhou, M, Grimshaw, S, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-05-20 | | Release date: | 2011-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.012 Å) | | Cite: | Crystal structure of a putative fumarylacetoacetate hydrolase family protein from Yersinia pestis CO92
TO BE PUBLISHED
|
|
3S83
 
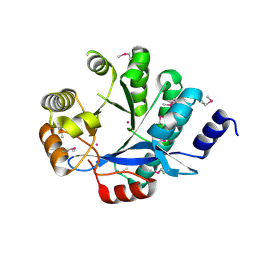 | | EAL domain of phosphodiesterase PdeA | | Descriptor: | GGDEF family protein, POTASSIUM ION | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Massa, C, Schirmer, T, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-05-27 | | Release date: | 2011-06-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal structure of EAL domain from Caulobacter crescentus CB15
To be Published
|
|
3SC6
 
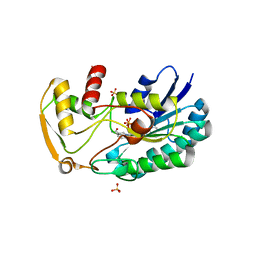 | | 2.65 Angstrom resolution crystal structure of dTDP-4-dehydrorhamnose reductase (rfbD) from Bacillus anthracis str. Ames in complex with NADP | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, SULFATE ION, dTDP-4-dehydrorhamnose reductase | | Authors: | Halavaty, A.S, Kuhn, M, Shuvalova, L, Minasov, G, Peterson, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-07 | | Release date: | 2011-06-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Bacillus anthracis dTDP-L-rhamnose-biosynthetic enzyme dTDP-4-dehydrorhamnose reductase (RfbD).
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3SD7
 
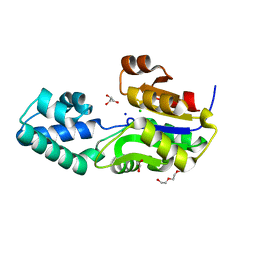 | | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative phosphatase, ... | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-06-08 | | Release date: | 2011-06-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 1.7 Angstrom Resolution Crystal Structure of Putative Phosphatase from Clostridium difficile.
TO BE PUBLISHED
|
|
5EZ4
 
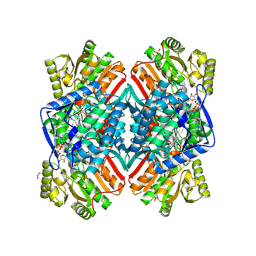 | | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-26 | | Release date: | 2015-12-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | 2.11 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M/Y450L double mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
5EYU
 
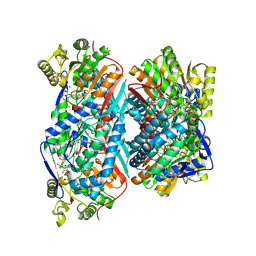 | | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Betaine aldehyde dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Halavaty, A.S, Minasov, G, Chen, C, Joo, J.C, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-11-25 | | Release date: | 2015-12-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | 1.72 Angstrom resolution crystal structure of betaine aldehyde dehydrogenase (betB) P449M point mutant from Staphylococcus aureus in complex with NAD+ and BME-modified Cys289
To Be Published
|
|
4YGO
 
 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae in intermediate state | | Descriptor: | CALCIUM ION, METHANOL, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Kiryukhina, O, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2015-02-26 | | Release date: | 2015-10-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
3HMQ
 
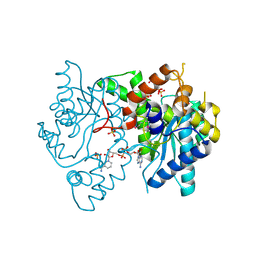 | | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+) | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Halavaty, A.S, Wawrzak, Z, Skarina, T, Onopriyenko, O, Peterson, S.N, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-05-29 | | Release date: | 2009-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom resolution crystal structure of a NAD synthetase (nadE) from Salmonella typhimurium LT2 in complex with NAD(+)
To be Published
|
|
3JZE
 
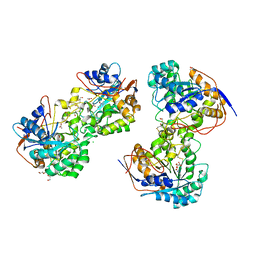 | | 1.8 Angstrom resolution crystal structure of dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2 | | Descriptor: | ACETIC ACID, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Minasov, G, Halavaty, A, Shuvalova, L, Dubrovska, I, Winsor, J, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-09-23 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 1.8 Angstrom Resolution Crystal Structure of Dihydroorotase (pyrC) from Salmonella enterica subsp. enterica serovar Typhimurium str. LT2.
TO BE PUBLISHED
|
|
3BV6
 
 | | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold | | Descriptor: | FE (III) ION, Metal-dependent hydrolase | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Yang, X, Collart, F.R, Joachimiak, A, Anderson, W.F, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-04 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of uncharacterized metallo protein from Vibrio cholerae with beta-lactamase like fold.
To be Published
|
|
3EDN
 
 | | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family | | Descriptor: | MAGNESIUM ION, Phenazine biosynthesis protein, PhzF family, ... | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-09-03 | | Release date: | 2008-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus anthracis phenazine biosynthesis protein, PhzF family
To be Published
|
|
3FF1
 
 | | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus | | Descriptor: | GLUCOSE-6-PHOSPHATE, Glucose-6-phosphate isomerase, SODIUM ION | | Authors: | Anderson, S.M, Brunzelle, J.S, Onopriyenko, O, Peterson, S, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2008-12-01 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Glucose 6-phosphate Isomerase from Staphylococcus aureus
TO BE PUBLISHED
|
|
3SS6
 
 | | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase | | Descriptor: | Acetyl-CoA acetyltransferase, POTASSIUM ION, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Peterson, S.N, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-07 | | Release date: | 2011-07-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Bacillus anthracis acetyl-CoA acetyltransferase
TO BE PUBLISHED
|
|
