3NKP
 
 | | Crystal structure of mouse autotaxin in complex with 18:1-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKR
 
 | | Crystal structure of mouse autotaxin in complex with 22:6-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (4Z,7E,10E,13Z,16Z,19Z)-docosa-4,7,10,13,16,19-hexaenoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.704 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKN
 
 | | Crystal structure of mouse autotaxin in complex with 14:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl tetradecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKO
 
 | | Crystal structure of mouse autotaxin in complex with 16:0-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl hexadecanoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKM
 
 | | Crystal structure of mouse autotaxin | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
3NKQ
 
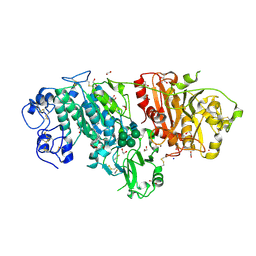 | | Crystal structure of mouse autotaxin in complex with 18:3-LPA | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E,12Z,15Z)-octadeca-9,12,15-trienoate, 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nishimasu, H, Ishitani, R, Mihara, E, Takagi, J, Aoki, J, Nureki, O. | | Deposit date: | 2010-06-20 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of autotaxin and insight into GPCR activation by lipid mediators
Nat.Struct.Mol.Biol., 18, 2011
|
|
7CEC
 
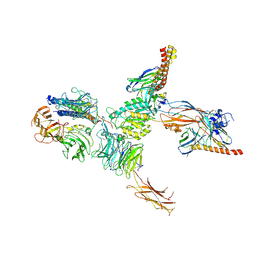 | | Structure of alpha6beta1 integrin in complex with laminin-511 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Arimori, T, Miyazaki, N, Takagi, J. | | Deposit date: | 2020-06-22 | | Release date: | 2021-06-23 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural mechanism of laminin recognition by integrin.
Nat Commun, 12, 2021
|
|
7CEA
 
 | |
7CEB
 
 | |
7DMU
 
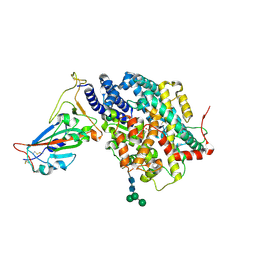 | |
2DDU
 
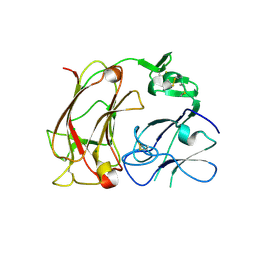 | | Crystal structure of the third repeat domain of reelin | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Nogi, T, Yasui, N, Takagi, J. | | Deposit date: | 2006-02-03 | | Release date: | 2006-09-26 | | Last modified: | 2011-10-05 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of a signaling-competent reelin fragment revealed by X-ray crystallography and electron tomography
Embo J., 25, 2006
|
|
4YO0
 
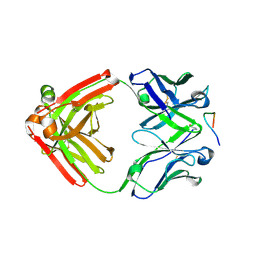 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 with bound PA peptide | | Descriptor: | 1,2-ETHANEDIOL, Heavy chain of antigen binding fragment, Fab, ... | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
4YNY
 
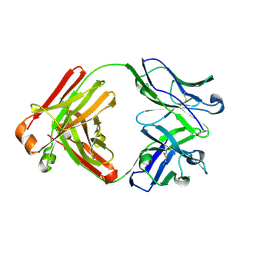 | | Crystal structure of monoclonal anti-human podoplanin antibody NZ-1 | | Descriptor: | Heavy chain of antigen binding fragment, Fab, Light chain of antigen binding fragment | | Authors: | Fujii, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2015-03-11 | | Release date: | 2016-03-02 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.584 Å) | | Cite: | Tailored placement of a turn-forming PA tag into the structured domain of a protein to probe its conformational state
J.Cell.Sci., 129, 2016
|
|
5B4Y
 
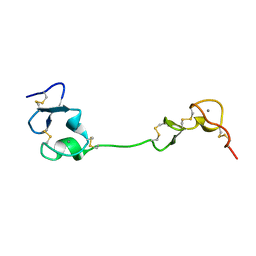 | | Crystal structure of the LA12 fragment of ApoER2 | | Descriptor: | CALCIUM ION, Low-density lipoprotein receptor-related protein 8 | | Authors: | Nogi, T, Tabata, S, Hirai, H, Yasui, N, Takagi, J. | | Deposit date: | 2016-04-20 | | Release date: | 2017-04-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
To Be Published
|
|
5AUM
 
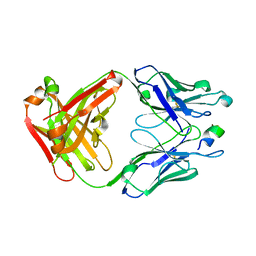 | |
5B4W
 
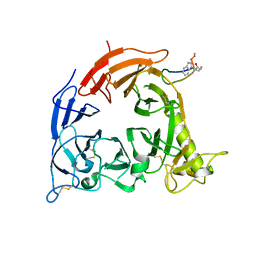 | | Crystal structure of Plexin inhibitor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-B1, Synthesized cyclic peptide | | Authors: | Matsunaga, Y, Kitago, Y, Arimori, T, Takagi, J. | | Deposit date: | 2016-04-19 | | Release date: | 2016-12-28 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Allosteric Inhibition of a Semaphorin 4D Receptor Plexin B1 by a High-Affinity Macrocyclic Peptide
Cell Chem Biol, 23, 2016
|
|
5B4X
 
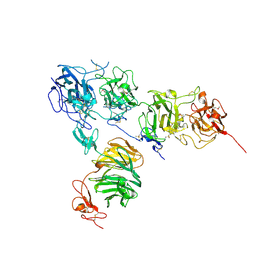 | | Crystal structure of the ApoER2 ectodomain in complex with the Reelin R56 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low density lipoprotein receptor-related protein 8, ... | | Authors: | Yasui, N, Hirai, H, Yamashita, K, Takagi, J, Nogi, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
To Be Published
|
|
6LZ4
 
 | |
5XCQ
 
 | | Crystal structure of P20.1 Fv-clasp fragment with its antigen peptide | | Descriptor: | C8 peptide, SULFATE ION, VH-SARAH(Y35C)chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.313 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCV
 
 | | Crystal structure of NZ-1 Fv-clasp fragment with its antigen peptide | | Descriptor: | CALCIUM ION, PA14 peptide, VH(S112C)-SARAH chimera,VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.143 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
5XCT
 
 | | Crystal structure of P20.1 Fv-clasp fragment with its antigen peptide | | Descriptor: | C8 peptide, CHLORIDE ION, VH(S112C)-SARAH chimera, ... | | Authors: | Arimori, T, Takagi, J. | | Deposit date: | 2017-03-23 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Fv-clasp: An Artificially Designed Small Antibody Fragment with Improved Production Compatibility, Stability, and Crystallizability
Structure, 25, 2017
|
|
7XOS
 
 | |
7XOL
 
 | |
7XOK
 
 | |
7XON
 
 | |
