1QZV
 
 | | Crystal structure of plant photosystem I | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHYLLOQUINONE, ... | | Authors: | Ben-Shem, A, Frolow, F, Nelson, N. | | Deposit date: | 2003-09-18 | | Release date: | 2004-01-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (4.44 Å) | | Cite: | Crystal structure of plant photosystem I.
Nature, 426, 2003
|
|
2HOX
 
 | | alliinase from allium sativum (garlic) | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Shimon, L.J.W, Rabinkov, A, Wilcheck, M, Mirelman, D, Frolow, F. | | Deposit date: | 2006-07-17 | | Release date: | 2007-02-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Two Structures of Alliinase from Alliium sativum L.: Apo Form and Ternary Complex with Aminoacrylate Reaction Intermediate Covalently Bound to the PLP Cofactor.
J.Mol.Biol., 366, 2007
|
|
3BWZ
 
 | | Crystal structure of the type II cohesin module from the cellulosome of Acetivibrio cellulolyticus with an extended linker conformation | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Cellulosomal scaffoldin adaptor protein B, ... | | Authors: | Noach, I, Lamed, R, Shimon, L.J.W, Bayer, E, Frolow, F. | | Deposit date: | 2008-01-10 | | Release date: | 2009-01-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
4IU3
 
 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, Cell-wall anchoring protein, Cellulose-binding protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
4IPY
 
 | | HIV capsid C-terminal domain | | Descriptor: | 1,2-ETHANEDIOL, Capsid protein p24 | | Authors: | Lampel, A, Yaniv, O, Berger, O, Bachrach, E, Gazit, E, Frolow, F. | | Deposit date: | 2013-01-10 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | A triclinic crystal structure of the carboxy-terminal domain of HIV-1 capsid protein with four molecules in the asymmetric unit reveals a novel packing interface.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4JO5
 
 | | CBM3a-L domain with flanking linkers from scaffoldin cipA of cellulosome of Clostridium thermocellum | | Descriptor: | 1-METHYLIMIDAZOLE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Shimon, L.J.W, Frolow, F, Bayer, E.A, Yaniv, O, Lamed, R, Morag, E. | | Deposit date: | 2013-03-16 | | Release date: | 2013-07-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structure of a family 3a carbohydrate-binding module from the cellulosomal scaffoldin CipA of Clostridium thermocellum with flanking linkers: implications for cellulosome structure.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
4IU2
 
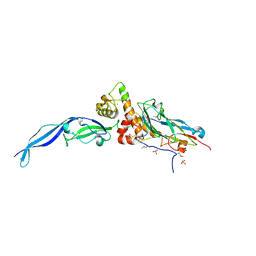 | | Cohesin-dockerin -X domain complex from Ruminococcus flavefacience | | Descriptor: | CALCIUM ION, CHLORIDE ION, Cell-wall anchoring protein, ... | | Authors: | Salama-Alber, O, Bayer, E, Frolow, F. | | Deposit date: | 2013-01-19 | | Release date: | 2013-04-24 | | Last modified: | 2013-07-03 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Atypical Cohesin-Dockerin Complex Responsible for Cell Surface Attachment of Cellulosomal Components: BINDING FIDELITY, PROMISCUITY, AND STRUCTURAL BUTTRESSES.
J.Biol.Chem., 288, 2013
|
|
1TYJ
 
 | | Crystal Structure Analysis of type II Cohesin A11 from Bacteroides cellulosolvens | | Descriptor: | 1,2-ETHANEDIOL, METHANOL, cellulosomal scaffoldin | | Authors: | Noach, I, Frolow, F, Jakoby, H, Rosenheck, S, Shimon, L.J.W, Lamed, R, Bayer, E.A. | | Deposit date: | 2004-07-08 | | Release date: | 2005-04-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements
J.Mol.Biol., 348, 2005
|
|
6Q61
 
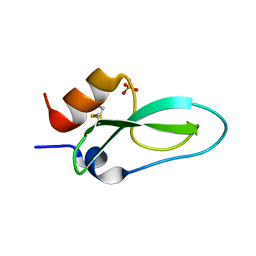 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | Kunitz-type conkunitzin-S1, SULFATE ION | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1YKF
 
 | | NADP-DEPENDENT ALCOHOL DEHYDROGENASE FROM THERMOANAEROBIUM BROCKII | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADP-DEPENDENT ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Korkhin, Y, Frolow, F. | | Deposit date: | 1996-03-25 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | NADP-dependent bacterial alcohol dehydrogenases: crystal structure, cofactor-binding and cofactor specificity of the ADHs of Clostridium beijerinckii and Thermoanaerobacter brockii.
J.Mol.Biol., 278, 1998
|
|
1ZV9
 
 | | Crystal structure analysis of a type II cohesin domain from the cellulosome of Acetivibrio cellulolyticus- SeMet derivative | | Descriptor: | 1,2-ETHANEDIOL, 1,3-PROPANDIOL, ACETIC ACID, ... | | Authors: | Noach, I, Rosenheck, S, Lamed, R, Shimon, L, Bayer, E, Frolow, F. | | Deposit date: | 2005-06-01 | | Release date: | 2006-06-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of Acetivibrio cellulolyticus.
J.Mol.Biol., 391, 2009
|
|
6Q6C
 
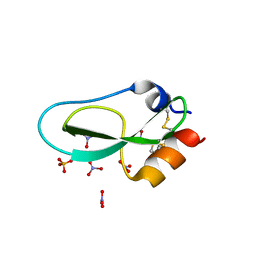 | | Pore-modulating toxins exploit inherent slow inactivation to block K+ channels | | Descriptor: | 1,2-ETHANEDIOL, Kunitz-type conkunitzin-S1, NITRATE ION, ... | | Authors: | Karbat, I, Gueta, H, Fine, S, Szanto, T, Hamer-Rogotner, S, Dym, O, Frolow, F, Gordon, D, Panyi, G, Gurevitz, M, Reuveny, E. | | Deposit date: | 2018-12-10 | | Release date: | 2019-08-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Pore-modulating toxins exploit inherent slow inactivation to block K+channels.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1Y9A
 
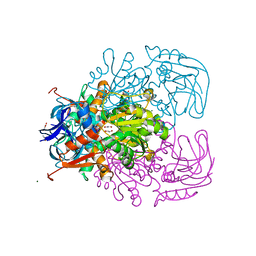 | | Alcohol Dehydrogenase from Entamoeba histolotica in complex with cacodylate | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CACODYLATE ION, ... | | Authors: | Shimon, L.J, Peretz, M, Goihberg, E, Burstein, Y, Frolow, F. | | Deposit date: | 2004-12-15 | | Release date: | 2006-01-17 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structure of alcohol dehydrogenase from Entamoeba histolytica.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1BFR
 
 | | IRON STORAGE AND ELECTRON TRANSPORT | | Descriptor: | BACTERIOFERRITIN, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Dautant, A, Yariv, J, Meyer, J.B, Precigoux, G, Sweet, R.M, Frolow, F, Kalb(Gilboa), A.J. | | Deposit date: | 1994-12-16 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Structure of a monoclinic crystal from of cyctochrome b1 (Bacterioferritin) from E. coli.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1D31
 
 | | THE THREE-DIMENSIONAL STRUCTURES OF BULGE-CONTAINING DNA FRAGMENTS | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Joshua-Tor, L, Frolow, F, Appella, E, Hope, H, Rabinovich, D, Sussman, J.L. | | Deposit date: | 1991-04-25 | | Release date: | 1992-04-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Three-dimensional structures of bulge-containing DNA fragments.
J.Mol.Biol., 225, 1992
|
|
2D94
 
 | | THE CONFORMATION OF THE DNA DOUBLE HELIX IN THE CRYSTAL IS DEPENDENT ON ITS ENVIRONMENT | | Descriptor: | DNA (5'-D(*GP*GP*GP*CP*GP*CP*CP*C)-3') | | Authors: | Shakked, Z, Guerstein-Guzikevich, G, Eisenstein, M, Frolow, F, Rabinovich, D. | | Deposit date: | 1993-07-13 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The conformation of the DNA double helix in the crystal is dependent on its environment.
Nature, 342, 1989
|
|
1PED
 
 | | BACTERIAL SECONDARY ALCOHOL DEHYDROGENASE (APO-FORM) | | Descriptor: | NADP-DEPENDENT ALCOHOL DEHYDROGENASE, ZINC ION | | Authors: | Korkhin, Y, Frolow, F. | | Deposit date: | 1995-12-28 | | Release date: | 1997-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystalline alcohol dehydrogenases from the mesophilic bacterium Clostridium beijerinckii and the thermophilic bacterium Thermoanaerobium brockii: preparation, characterization and molecular symmetry.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1LLU
 
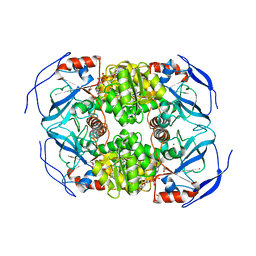 | | THE TERNARY COMPLEX OF PSEUDOMONAS AERUGINOSA ALCOHOL DEHYDROGENASE WITH ITS COENZYME AND WEAK SUBSTRATE | | Descriptor: | 1,2-ETHANEDIOL, Alcohol Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Levin, I, Meiri, G, Peretz, M, Frolow, F, Burstein, Y. | | Deposit date: | 2002-04-30 | | Release date: | 2003-11-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The ternary complex of Pseudomonas aeruginosa alcohol dehydrogenase with NADH and ethylene glycol.
Protein Sci., 13, 2004
|
|
1LJM
 
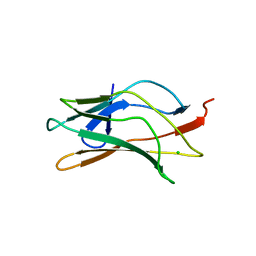 | | DNA recognition is mediated by conformational transition and by DNA bending | | Descriptor: | CHLORIDE ION, RUNX1 transcription factor | | Authors: | Bartfeld, D, Shimon, L, Couture, G.C, Rabinovich, D, Frolow, F, Levanon, D, Groner, Y, Shakked, Z. | | Deposit date: | 2002-04-22 | | Release date: | 2002-11-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | DNA Recognition by the RUNX1 Transcription Factor Is Mediated by an Allosteric Transition in the RUNT Domain and by DNA Bending.
Structure, 10
|
|
1KEV
 
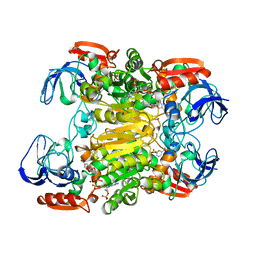 | | STRUCTURE OF NADP-DEPENDENT ALCOHOL DEHYDROGENASE | | Descriptor: | NADP-DEPENDENT ALCOHOL DEHYDROGENASE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ZINC ION | | Authors: | Korkhin, Y, Frolow, F. | | Deposit date: | 1996-10-21 | | Release date: | 1997-10-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystalline alcohol dehydrogenases from the mesophilic bacterium Clostridium beijerinckii and the thermophilic bacterium Thermoanaerobium brockii: preparation, characterization and molecular symmetry.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
1JQB
 
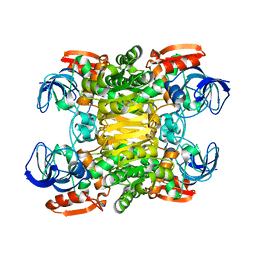 | | Alcohol Dehydrogenase from Clostridium Beijerinckii: Crystal Structure of Mutant with Enhanced Thermal Stability | | Descriptor: | NADP-dependent Alcohol Dehydrogenase, ZINC ION | | Authors: | Levin, I, Frolow, F, Bogin, O, Peretz, M, Hacham, Y, Burstein, Y. | | Deposit date: | 2001-08-05 | | Release date: | 2002-11-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural basis for the enhanced thermal stability of alcohol dehydrogenase
mutants from the mesophilic bacterium Clostridium beijerinckii:
contribution of salt bridging
Protein Sci., 11, 2002
|
|
1G43
 
 | | CRYSTAL STRUCTURE OF A FAMILY IIIA CBD FROM CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | CALCIUM ION, SCAFFOLDING PROTEIN, ZINC ION | | Authors: | Shimon, L.J.W, Pages, S, Belaich, A, Belaich, J.-P, Bayer, E.A, Lamed, R, Shoham, Y, Frolow, F. | | Deposit date: | 2000-10-26 | | Release date: | 2000-12-01 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of a family IIIa scaffoldin CBD from the cellulosome of Clostridium cellulolyticum at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2YLK
 
 | | Carbohydrate-binding module CBM3b from the cellulosomal cellobiohydrolase 9A from Clostridium thermocellum | | Descriptor: | CELLULOSE 1,4-BETA-CELLOBIOSIDASE | | Authors: | Yaniv, O, Petkun, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2011-06-02 | | Release date: | 2012-04-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A Single Mutation Reforms the Binding Activity of an Adhesion-Deficient Family 3 Carbohydrate-Binding Module
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2WNX
 
 | | 3b' carbohydrate-binding module from the Cel9V glycoside hydrolase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, FORMIC ACID, GLYCOSIDE HYDROLASE, ... | | Authors: | Petkun, S, Jindou, S, Shimon, L.J.W, Bayer, E.A, Lamed, R, Frolow, F. | | Deposit date: | 2009-07-20 | | Release date: | 2009-12-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure of a Family 3B' Carbohydrate-Binding Module from the Cel9V Glycoside Hydrolase from Clostridium Thermocellum: Structural Diversity and Implications for Carbohydrate Binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2HOR
 
 | | Crystal structure of alliinase from garlic- apo form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Alliin lyase 1, ... | | Authors: | Shimon, L.J.W, Rabinkov, A, Wilcheck, M, Mirelman, D, Frolow, F. | | Deposit date: | 2006-07-16 | | Release date: | 2007-02-06 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Two Structures of Alliinase from Alliium sativum L.: Apo Form and Ternary Complex with Aminoacrylate Reaction Intermediate Covalently Bound to the PLP Cofactor.
J.Mol.Biol., 366, 2007
|
|
