4MEX
 
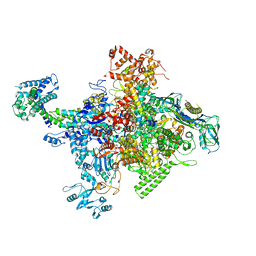 | | Crystal structure of Escherichia coli RNA polymerase in complex with salinamide A | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Feng, Y, Zhang, Y, Arnold, E, Ebright, R.H. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-21 | | Last modified: | 2014-06-25 | | Method: | X-RAY DIFFRACTION (3.902 Å) | | Cite: | Transcription inhibition by the depsipeptide antibiotic salinamide A.
Elife, 3, 2014
|
|
4E96
 
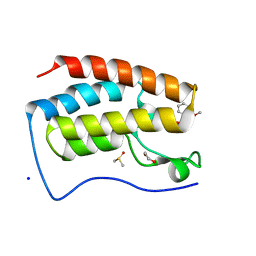 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor PFi-1 | | Descriptor: | 1,2-ETHANEDIOL, 2-methoxy-N-(3-methyl-2-oxo-1,2,3,4-tetrahydroquinazolin-6-yl)benzenesulfonamide, Bromodomain-containing protein 4, ... | | Authors: | Filippakopoulos, P, Picaud, S, Felletar, I, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Fish, P, Bunnage, M, Owen, D, Knapp, S, Cook, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-03-20 | | Release date: | 2012-04-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a chemical probe for bromo and extra C-terminal bromodomain inhibition through optimization of a fragment-derived hit.
J.Med.Chem., 55, 2012
|
|
1PER
 
 | |
4AXL
 
 | | HUMAN CATHEPSIN L APO FORM WITH ZN | | Descriptor: | ACETATE ION, CATHEPSIN L1, GLYCEROL, ... | | Authors: | Banner, D.W, Benz, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Optimization of Triazine Nitriles as Rhodesain Inhibitors: Structure-Activity Relationships, Bioisosteric Imidazopyridine Nitriles, and X-Ray Crystal Structure Analysis with Human Cathepsin L
Chemmedchem, 8, 2013
|
|
3T03
 
 | | Crystal structure of PPAR gamma ligand binding domain in complex with a novel partial agonist GQ-16 | | Descriptor: | (5Z)-5-(5-bromo-2-methoxybenzylidene)-3-(4-methylbenzyl)-1,3-thiazolidine-2,4-dione, Nuclear receptor coactivator 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Rajagopalan, S, Webb, P, Baxter, J.D, Brennan, R.G, Phillips, K.J. | | Deposit date: | 2011-07-19 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | GQ-16, a novel peroxisome proliferator-activated receptor (PPAR gamma) ligand, promotes insulin sensitization without weight gain.
J.Biol.Chem., 287, 2012
|
|
4AXM
 
 | | TRIAZINE CATHEPSIN INHIBITOR COMPLEX | | Descriptor: | 4-[(4-chlorobenzyl)(cyclohexyl)amino]-6-morpholin-4-yl-1,3,5-triazine-2-carboxamide, CATHEPSIN L1, GLYCEROL | | Authors: | Ehmke, V, Diederich, F, Banner, D.W, Benz, J. | | Deposit date: | 2012-06-13 | | Release date: | 2013-05-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Optimization of Triazine Nitriles as Rhodesain Inhibitors: Structure-Activity Relationships, Bioisosteric Imidazopyridine Nitriles, and X-Ray Crystal Structure Analysis with Human Cathepsin L
Chemmedchem, 8, 2013
|
|
4J23
 
 | | Low resolution crystal structure of the FGFR2D2D3/FGF1/SR128545 complex | | Descriptor: | Fibroblast growth factor 1, Fibroblast growth factor receptor 2 | | Authors: | Kudlinzki, D, Saxena, K, Sreeramulu, S, Schieborr, U, Dreyer, M, Schreuder, H, Schwalbe, H. | | Deposit date: | 2013-02-04 | | Release date: | 2014-02-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.882 Å) | | Cite: | Molecular mechanism of SSR128129E, an extracellularly acting, small-molecule, allosteric inhibitor of FGF receptor signaling.
Cancer Cell, 23, 2013
|
|
4BYY
 
 | | Apo GlxR | | Descriptor: | GLYCEROL, PHOSPHATE ION, TRANSCRIPTIONAL REGULATOR, ... | | Authors: | Pohl, E, Cann, M.J, Townsend, P.D, Money, V.A. | | Deposit date: | 2013-07-21 | | Release date: | 2014-07-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | The Crystal Structures of Apo and Camp-Bound Glxr from Corynebacterium Glutamicum Reveal Structural and Dynamic Changes Upon Camp Binding in Crp/Fnr Family Transcription Factors.
Plos One, 9, 2014
|
|
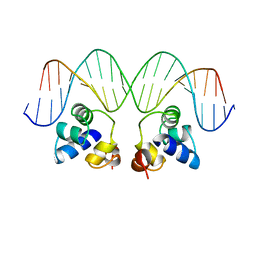
3CRO
 
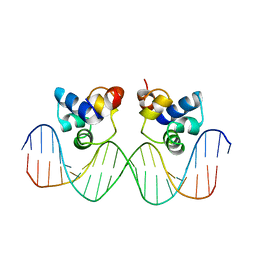 | | THE PHAGE 434 CRO/OR1 COMPLEX AT 2.5 ANGSTROMS RESOLUTION | | Descriptor: | DNA (5'-D(*AP*AP*GP*TP*AP*CP*AP*AP*AP*CP*TP*TP*TP*CP*TP*TP*G P*TP*AP*T)-3'), DNA (5'-D(*TP*AP*TP*AP*CP*AP*AP*GP*AP*AP*AP*GP*TP*TP*TP*GP*T P*AP*CP*T)-3'), PROTEIN (434 CRO) | | Authors: | Mondragon, A, Harrison, S.C. | | Deposit date: | 1990-07-06 | | Release date: | 1991-10-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The phage 434 Cro/OR1 complex at 2.5 A resolution.
J.Mol.Biol., 219, 1991
|
|
6UOG
 
 | | Asparaginase II from Escherichia coli | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Araujo, T.S, Almeida, M.S, Lima, L.M.T.R. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Biophysical characterization of two commercially available preparations of the drug containing Escherichia coli L-Asparaginase 2.
Biophys.Chem., 271, 2021
|
|
5XMR
 
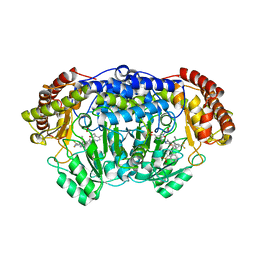 | | Plasmodium vivax SHMT(C346A) bound with PLP-glycine and GS395 | | Descriptor: | (4~{S})-6-azanyl-3-methyl-4-[3-[4-(phenylmethyl)sulfonylphenyl]-5-(trifluoromethyl)phenyl]-4-propan-2-yl-2~{H}-pyrano[2 ,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
6UOD
 
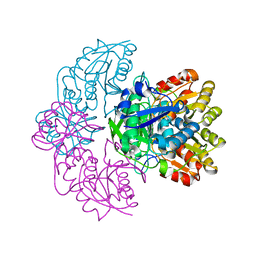 | | Asparaginase II from Escherichia coli | | Descriptor: | L-asparaginase 2 | | Authors: | Araujo, T.S, Almeida, M.S, Lima, L.M.T.R. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Biophysical characterization of two commercially available preparations of the drug containing Escherichia coli L-Asparaginase 2.
Biophys.Chem., 271, 2021
|
|
6UOH
 
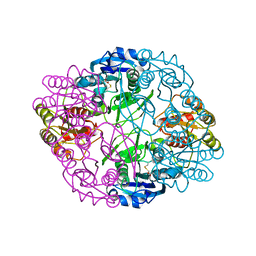 | | Asparaginase II from Escherichia coli | | Descriptor: | ASPARTIC ACID, L-asparaginase 2 | | Authors: | Araujo, T.S, Almeida, M.S, Lima, L.M.T.R. | | Deposit date: | 2019-10-14 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biophysical characterization of two commercially available preparations of the drug containing Escherichia coli L-Asparaginase 2.
Biophys.Chem., 271, 2021
|
|
6V3V
 
 | |
5YG4
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and S-GS849 | | Descriptor: | 2-[1-[(3~{S})-6'-azanyl-5'-cyano-7-fluoranyl-2,2,3'-trimethyl-spiro[1~{H}-indene-3,4'-2~{H}-pyrano[2,3-c]pyrazole]-5-yl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
8GLV
 
 | |
5YG2
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS705 | | Descriptor: | 3-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidin-4-yl]propanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
6VAS
 
 | |
5YG3
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and S-GS834 | | Descriptor: | (3~{S})-6'-azanyl-7-fluoranyl-2,2,3'-trimethyl-5-pyridin-4-yl-spiro[1~{H}-indene-3,4'-2~{H}-pyrano[2,3-c]pyrazole]-5'-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
5YFZ
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and S-GS626 | | Descriptor: | 2-[1-[3-[(4~{S})-6-azanyl-5-cyano-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazol-4-yl]-5-fluoranyl-phenyl]piperidin-4-yl]ethanoic acid, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-09-22 | | Release date: | 2018-06-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Potent Inhibitors of Plasmodial Serine Hydroxymethyltransferase (SHMT) Featuring a Spirocyclic Scaffold
ChemMedChem, 13, 2018
|
|
5XMV
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS362 | | Descriptor: | (4~{S})-6-azanyl-4-[3-(2-chlorophenyl)-5-(trifluoromethyl)phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5- carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5XMT
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS380 | | Descriptor: | (4~{S})-6-azanyl-4-[3-(2-cyanophenyl)-5-(trifluoromethyl)phenyl]-3-methyl-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
5XMU
 
 | | Plasmodium vivax SHMT bound with PLP-glycine and GS363 | | Descriptor: | (4~{S})-6-azanyl-3-methyl-4-[3-(2-methylphenyl)-5-(trifluoromethyl)phenyl]-4-propan-2-yl-2~{H}-pyrano[2,3-c]pyrazole-5-carbonitrile, CHLORIDE ION, N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], ... | | Authors: | Chitnumsub, P, Jaruwat, A, Leartsakulpanich, U, Schwertz, G, Diederich, F. | | Deposit date: | 2017-05-16 | | Release date: | 2017-11-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Conformational Aspects in the Design of Inhibitors for Serine Hydroxymethyltransferase (SHMT): Biphenyl, Aryl Sulfonamide, and Aryl Sulfone Motifs
Chemistry, 23, 2017
|
|
1QNK
 
 | | TRUNCATED HUMAN GROB[5-73], NMR, 20 STRUCTURES | | Descriptor: | C-X-C motif chemokine 2 | | Authors: | Qian, Y.Q, Johanson, K, McDevitt, P. | | Deposit date: | 1999-10-18 | | Release date: | 2000-02-04 | | Last modified: | 2018-06-13 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution structure of truncated human GRObeta [5-73] and its structural comparison with CXC chemokine family members GROalpha and IL-8.
J. Mol. Biol., 294, 1999
|
|
6VJO
 
 | |
