1FP7
 
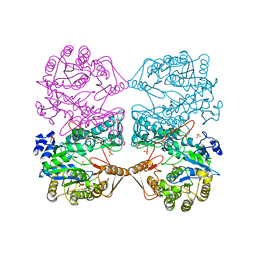 | | MONOVALENT CATION BINDING SITES IN N10-FORMYLTETRAHYDROFOLATE SYNTHETASE FROM MOORELLA THERMOACETICA | | Descriptor: | FORMATE--TETRAHYDROFOLATE LIGASE, POTASSIUM ION, SULFATE ION | | Authors: | Radfar, R, Leaphart, A, Brewer, J.M, Minor, W, Odom, J.D. | | Deposit date: | 2000-08-30 | | Release date: | 2001-08-30 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cation binding and thermostability of FTHFS monovalent cation binding sites and thermostability of N10-formyltetrahydrofolate synthetase from Moorella thermoacetica.
Biochemistry, 39, 2000
|
|
6TTO
 
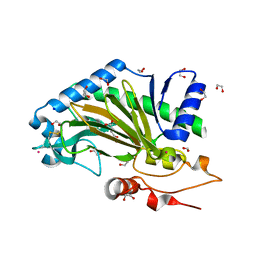 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with 2-oxoglutarate | | Descriptor: | 1,2-ETHANEDIOL, 2-OXOGLUTARIC ACID, FORMIC ACID, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
6TTN
 
 | | N-terminally truncated hyoscyamine 6-hydroxylase (tH6H) in complex with N-oxalylglycine and hyoscyamine | | Descriptor: | 1,2-ETHANEDIOL, Hyoscyamine 6 beta-hydroxylase, N-OXALYLGLYCINE, ... | | Authors: | Kluza, A, Mrugala, B, Porebski, P.J, Kurpiewska, K, Niedzialkowska, E, Weiss, M.S, Borowski, T. | | Deposit date: | 2019-12-29 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | Regioselectivity of hyoscyamine 6 beta-hydroxylase-catalysed hydroxylation as revealed by high-resolution structural information and QM/MM calculations.
Dalton Trans, 49, 2020
|
|
5CNA
 
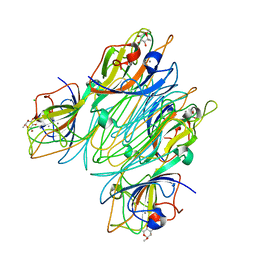 | | REFINED STRUCTURE OF CONCANAVALIN A COMPLEXED WITH ALPHA-METHYL-D-MANNOPYRANOSIDE AT 2.0 ANGSTROMS RESOLUTION AND COMPARISON WITH THE SACCHARIDE-FREE STRUCTURE | | Descriptor: | CALCIUM ION, CHLORIDE ION, CONCANAVALIN A, ... | | Authors: | Naismith, J.H, Emmerich, C, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Raftery, J, Kalb(Gilboa), A.J, Yariv, J. | | Deposit date: | 1994-02-11 | | Release date: | 1994-05-31 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of concanavalin A complexed with methyl alpha-D-mannopyranoside at 2.0 A resolution and comparison with the saccharide-free structure.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
8HLT
 
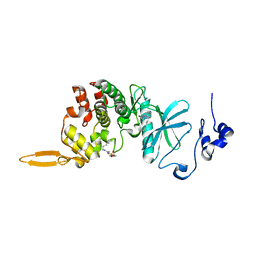 | | The co-crystal structure of DYRK2 with YK-2-99B | | Descriptor: | (6-{[(4P)-4-(1,3-benzothiazol-5-yl)-5-fluoropyrimidin-2-yl]amino}pyridin-3-yl)(piperazin-1-yl)methanone, Dual specificity tyrosine-phosphorylation-regulated kinase 2 | | Authors: | Shen, H.T, Xiao, Y.B, Yuan, K, Yang, P, Li, Q.N. | | Deposit date: | 2022-12-01 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of Potent DYRK2 Inhibitors with High Selectivity, Great Solubility, and Excellent Safety Properties for the Treatment of Prostate Cancer.
J.Med.Chem., 66, 2023
|
|
3RQF
 
 | |
3RQG
 
 | |
3RQE
 
 | |
7VUN
 
 | | Design, modification, evaluation and cocrystal studies of novel phthalimides regulating PD-1/PD-L1 interaction | | Descriptor: | (2~{S},3~{S})-2-[[6-[(3-cyanophenyl)methoxy]-2-(2-methyl-3-phenyl-phenyl)-1,3-bis(oxidanylidene)isoindol-5-yl]methylamino]-3-oxidanyl-butanoic acid, Programmed cell death 1 ligand 1 | | Authors: | Cheng, Y, Sun, C.L, Chen, M.R, Yang, P, Xiao, Y.B. | | Deposit date: | 2021-11-03 | | Release date: | 2022-09-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.701 Å) | | Cite: | Novel phthalimides regulating PD-1/PD-L1 interaction as potential immunotherapy agents.
Acta Pharm Sin B, 12, 2022
|
|
7EJV
 
 | | The co-crystal structure of DYRK2 with YK-2-69 | | Descriptor: | Dual specificity tyrosine-phosphorylation-regulated kinase 2, [6-[[4-[2-(dimethylamino)-1,3-benzothiazol-6-yl]-5-fluoranyl-pyrimidin-2-yl]amino]pyridin-3-yl]-(4-ethylpiperazin-1-yl)methanone | | Authors: | Li, Z, Xiao, Y, Yuan, K, Kuang, W, Xiuquan, Y, Yang, P. | | Deposit date: | 2021-04-02 | | Release date: | 2022-04-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting dual-specificity tyrosine phosphorylation-regulated kinase 2 with a highly selective inhibitor for the treatment of prostate cancer.
Nat Commun, 13, 2022
|
|
6OV8
 
 | | 2.6 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Peptidase B, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-05-07 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
6OAD
 
 | | 2.05 Angstrom Resolution Crystal Structure of Aminopeptidase B from Escherichia coli str. K-12 substr. MG1655. | | Descriptor: | 1,2-ETHANEDIOL, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Minasov, G, Shuvalova, L, Wawrzak, Z, Kiryukhina, O, Grimshaw, S, Kwon, K, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-03-15 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparison of metal-bound and unbound structures of aminopeptidase B proteins from Escherichia coli and Yersinia pestis.
Protein Sci., 29, 2020
|
|
1SCS
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, COBALT (II) ION, CONCANAVALIN A | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1SCR
 
 | | HIGH-RESOLUTION STRUCTURES OF SINGLE-METAL-SUBSTITUTED CONCANAVALIN A: THE CO,CA-PROTEIN AT 1.6 ANGSTROMS AND THE NI,CA-PROTEIN AT 2.0 ANGSTROMS | | Descriptor: | CALCIUM ION, CONCANAVALIN A, NICKEL (II) ION | | Authors: | Emmerich, C, Helliwell, J.R, Redshaw, M, Naismith, J.H, Harrop, S.J, Raftery, J, Kalb, A.J, Yariv, J, Dauter, Z, Wilson, K.S. | | Deposit date: | 1993-12-06 | | Release date: | 1994-05-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | High-resolution structures of single-metal-substituted concanavalin A: the Co,Ca-protein at 1.6 A and the Ni,Ca-protein at 2.0 A.
Acta Crystallogr.,Sect.D, 50, 1994
|
|
1JBC
 
 | | CONCANAVALIN A | | Descriptor: | CALCIUM ION, CONCANAVALIN A, MANGANESE (II) ION | | Authors: | Parkin, S, Rupp, B, Hope, H. | | Deposit date: | 1996-08-23 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Atomic resolution structure of concanavalin A at 120 K.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2CLQ
 
 | | Structure of mitogen-activated protein kinase kinase kinase 5 | | Descriptor: | MITOGEN-ACTIVATED PROTEIN KINASE KINASE KINASE 5, STAUROSPORINE | | Authors: | Bunkoczi, G, Salah, E, Fedorov, O, Pike, A, Gileadi, O, von Delft, F, Arrowsmith, C, Edwards, A, Sundstrom, M, Weigelt, J, Knapp, S. | | Deposit date: | 2006-04-28 | | Release date: | 2006-05-09 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Functional Characterization of the Human Protein Kinase Ask1.
Structure, 15, 2007
|
|
7P6F
 
 | | 1.93 A resolution X-ray crystal structure of the transcriptional regulator SrnR from Streptomyces griseus | | Descriptor: | ACETATE ION, SODIUM ION, Transcriptional regulator SrnR | | Authors: | Mazzei, L, Ciurli, S. | | Deposit date: | 2021-07-16 | | Release date: | 2022-05-25 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structure, dynamics, and function of SrnR, a transcription factor for nickel-dependent gene expression.
Metallomics, 13, 2021
|
|
1CON
 
 | | THE REFINED STRUCTURE OF CADMIUM SUBSTITUTED CONCANAVALIN A AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CADMIUM ION, CALCIUM ION, CONCANAVALIN A | | Authors: | Naismith, J.H, Habash, J, Harrop, S.J, Helliwell, J.R, Hunter, W.N, Kalb(Gilboa), A.J, Yariv, J, Wan, T.C.M, Weisgerber, S. | | Deposit date: | 1993-03-16 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined structure of cadmium-substituted concanavalin A at 2.0 A resolution.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
2CTV
 
 | |
3IRC
 
 | | Crystal structure analysis of dengue-1 envelope protein domain III | | Descriptor: | ENVELOPE PROTEIN, SULFATE ION | | Authors: | Nelson, C.A, Kim, T, Warren, J.T, Chruszcz, M, Minor, W, Fremont, D.H, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-21 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure Analysis of the Dengue-1 Envelope Protein Domain III
To be Published
|
|
7Q19
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W) in complex with desipramine (FAW-DSM#3) | | Descriptor: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | Authors: | Loch, J.I, Barciszewski, J, Pokrywka, K, Lewinski, K. | | Deposit date: | 2021-10-18 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7Q2N
 
 | | Beta-lactoglobulin mutant FAF (I56F/L39A/M107F) in complex with desipramine (FAF-DSM) | | Descriptor: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | Authors: | Loch, J.I, Barciszewski, J, Lewinski, K. | | Deposit date: | 2021-10-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7Q2P
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W) in complex with desipramine (FAW-DSM#2) | | Descriptor: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | Authors: | Loch, J.I, Barciszewski, J, Pokrywka, K, Lewinski, K. | | Deposit date: | 2021-10-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7Q17
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W), unliganded form | | Descriptor: | 1,2-ETHANEDIOL, Beta-lactoglobulin | | Authors: | Loch, J.I, Barciszewski, J, Lewinski, K. | | Deposit date: | 2021-10-18 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
7Q2O
 
 | | Beta-lactoglobulin mutant FAW (I56F/L39A/M107W) in complex with desipramine (FAW-DSM#1) | | Descriptor: | 1,2-ETHANEDIOL, 3-(10,11-DIHYDRO-5H-DIBENZO[B,F]AZEPIN-5-YL)-N-METHYLPROPAN-1-AMINE, Beta-lactoglobulin, ... | | Authors: | Loch, J.I, Barciszewski, J, Lewinski, K. | | Deposit date: | 2021-10-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | New ligand-binding sites identified in the crystal structures of [beta]-lactoglobulin complexes with desipramine
Iucrj, 9, 2022
|
|
