1B6U
 
 | |
1B8K
 
 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
2UWI
 
 | | Structure of CrmE, a poxvirus TNF receptor | | Descriptor: | CRME PROTEIN | | Authors: | Graham, S.C, Bahar, M.W, Abrescia, N.G, Smith, G.L, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2007-03-22 | | Release date: | 2007-07-10 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of Crme, a Virus-Encoded Tumour Necrosis Factor Receptor.
J.Mol.Biol., 372, 2007
|
|
2UXE
 
 | | The structure of Vaccinia virus N1 | | Descriptor: | HYPOTHETICAL PROTEIN | | Authors: | Cooray, S, Bahar, M.W, Abrescia, N.G.A, McVey, C.E, Bartlett, N.W, Chen, R.A.-J, Stuart, D.I, Grimes, J.M, Smith, G.L. | | Deposit date: | 2007-03-28 | | Release date: | 2007-05-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Functional and Structural Studies of the Vaccinia Virus Virulence Factor N1 Reveal a Bcl-2-Like Anti- Apoptotic Protein
J.Gen.Virol., 88, 2007
|
|
2UUX
 
 | | Structure of the tryptase inhibitor TdPI from a tick | | Descriptor: | SULFATE ION, TRYPTASE INHIBITOR | | Authors: | Siebold, C, Paesen, G.C, Harlos, K, Peacey, M.F, Nuttall, P.A, Stuart, D.I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-03 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | A Tick Protein with a Modified Kunitz Fold Inhibits Human Tryptase.
J.Mol.Biol., 368, 2007
|
|
2UUY
 
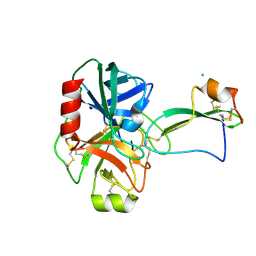 | | Structure of a tick tryptase inhibitor in complex with bovine trypsin | | Descriptor: | CALCIUM ION, CATIONIC TRYPSIN, CHLORIDE ION, ... | | Authors: | Siebold, C, Paesen, G.C, Harlos, K, Peacey, M.F, Nuttall, P.A, Stuart, D.I. | | Deposit date: | 2007-03-08 | | Release date: | 2007-04-10 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | A Tick Protein with a Modified Kunitz Fold Inhibits Human Tryptase.
J.Mol.Biol., 368, 2007
|
|
2VHC
 
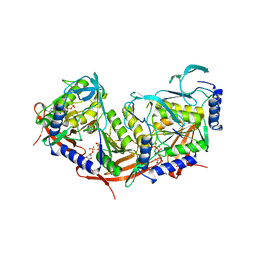 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 N234G mutant in complex with AMPCPP and MN | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MANGANESE (II) ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-20 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VD9
 
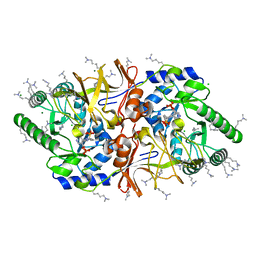 | | The crystal structure of alanine racemase from Bacillus anthracis (BA0252) with bound L-Ala-P | | Descriptor: | (1S)-1-[((1E)-{3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYLENE)AMINO]ETHYLPHOSPHONIC ACID, ALANINE RACEMASE, CHLORIDE ION, ... | | Authors: | Au, K, Ren, J, Walter, T.S, Harlos, K, Nettleship, J.E, Owens, R.J, Stuart, D.I, Esnouf, R.M, Oxford Protein Production Facility (OPPF), Structural Proteomics in Europe (SPINE) | | Deposit date: | 2007-10-01 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of an Alanine Racemase from Bacillus Anthracis (Ba0252) in the Presence and Absence of (R)-1-Aminoethylphosphonic Acid (L-Ala-P).
Acta Crystallogr.,Sect.F, 64, 2008
|
|
2WFF
 
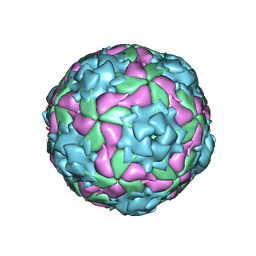 | | Equine Rhinitis A Virus | | Descriptor: | P1 | | Authors: | Fry, E.E, Tuthill, T.J, Harlos, K, Walter, T.S, Knowles, N.J, Gropelli, E, Rowlands, D.J, Stuart, D.I. | | Deposit date: | 2009-04-05 | | Release date: | 2010-05-05 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Equine Rhinitis a Virus and its Low Ph Empty Particle: Clues Towards an Aphthovirus Entry Mechanism?
Plos Pathog., 5, 2009
|
|
2VSM
 
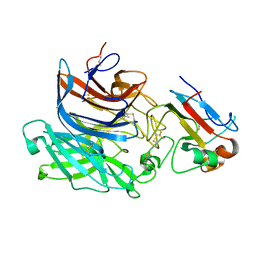 | | Nipah virus attachment glycoprotein in complex with human cell surface receptor ephrinB2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EPHRIN-B2, HEMAGGLUTININ-NEURAMINIDASE, ... | | Authors: | Bowden, T.A, Aricescu, A.R, Gilbert, R.J, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2008-04-25 | | Release date: | 2008-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Nipah and Hendra Virus Attachment to Their Cell-Surface Receptor Ephrin-B2
Nat.Struct.Mol.Biol., 15, 2008
|
|
2VVD
 
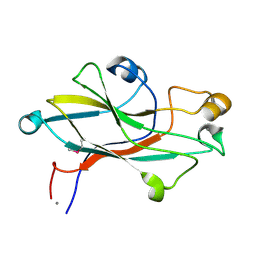 | | Crystal structure of the receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2W2R
 
 | | Structure of the vesicular stomatitis virus matrix protein | | Descriptor: | MATRIX PROTEIN | | Authors: | Graham, S.C, Assenberg, R, Delmas, O, Verma, A, Gholami, A, Talbi, C, Owens, R.J, Stuart, D.I, Grimes, J.M, Bourhy, H. | | Deposit date: | 2008-11-03 | | Release date: | 2009-01-13 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Rhabdovirus Matrix Protein Structures Reveal a Novel Mode of Self-Association.
Plos Pathog., 4, 2008
|
|
2VSC
 
 | | Structure of the immunoglobulin-superfamily ectodomain of human CD47 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LEUKOCYTE SURFACE ANTIGEN CD47, MAGNESIUM ION | | Authors: | Hatherley, D, Graham, S.C, Turner, J, Harlos, K, Stuart, D.I, Barclay, A.N. | | Deposit date: | 2008-04-22 | | Release date: | 2008-08-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Paired Receptor Specificity Explained by Structures of Signal Regulatory Proteins Alone and Complexed with Cd47.
Mol.Cell, 31, 2008
|
|
2VVY
 
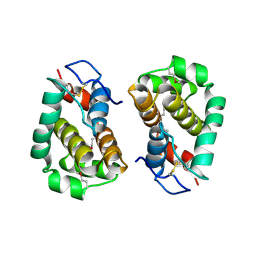 | | Structure of Vaccinia virus protein B14 | | Descriptor: | PROTEIN B15 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2019-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2VWD
 
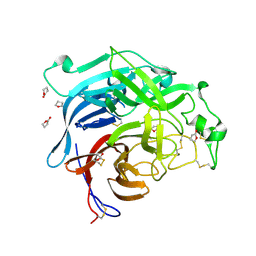 | | Nipah Virus Attachment Glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GAMMA-BUTYROLACTONE, ... | | Authors: | Bowden, T.A, Crispin, M, Harvey, D.J, Aricescu, A.R, Grimes, J.M, Jones, E.Y, Stuart, D.I. | | Deposit date: | 2008-06-20 | | Release date: | 2008-10-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structure and Carbohydrate Analysis of Nipah Virus Attachment Glycoprotein: A Template for Antiviral and Vaccine Design.
J.Virol., 82, 2008
|
|
2WAH
 
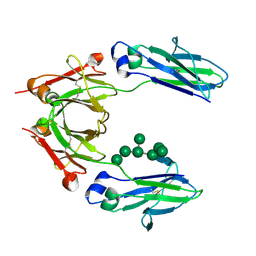 | | Crystal Structure of an IgG1 Fc Glycoform (Man9GlcNAc2) | | Descriptor: | IG GAMMA-1 CHAIN C REGION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Crispin, M, Bowden, T.A, Coles, C.H, Harlos, K, Aricescu, A.R, Harvey, D.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Carbohydrate and Domain Architecture of an Immature Antibody Glycoform Exhibiting Enhanced Effector Functions
J.Mol.Biol., 387, 2009
|
|
2WB1
 
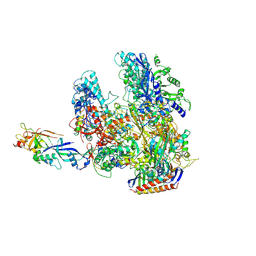 | | The complete structure of the archaeal 13-subunit DNA-directed RNA Polymerase | | Descriptor: | DNA-DIRECTED RNA POLYMERASE RPO10 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO11 SUBUNIT, DNA-DIRECTED RNA POLYMERASE RPO12 SUBUNIT, ... | | Authors: | Korkhin, Y, Unligil, U.M, Littlefield, O, Nelson, P.J, Stuart, D.I, Sigler, P.B, Bell, S.D, Abrescia, N.G.A. | | Deposit date: | 2009-02-19 | | Release date: | 2009-05-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.52 Å) | | Cite: | Evolution of Complex RNA Polymerase: The Complete Archaeal RNA Polymerase Structure
Plos Biol., 7, 2009
|
|
2W8X
 
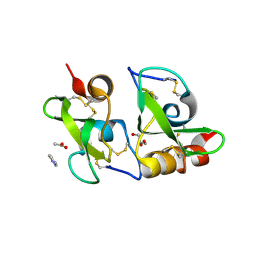 | | Structure of the tick ion-channel modulator Ra-KLP | | Descriptor: | ACETATE ION, GLYCEROL, IMIDAZOLE, ... | | Authors: | Paesen, G.C, Siebold, C, Dallas, M, Peers, C, Harlos, K, Nuttall, P.A, Nunn, M.A, Stuart, D.I, Esnouf, R.M. | | Deposit date: | 2009-01-20 | | Release date: | 2009-05-05 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An Ion-Channel Modulator from the Saliva of the Brown Ear Tick Has a Highly Modified Kunitz/Bpti Structure.
J.Mol.Biol., 389, 2009
|
|
2VVE
 
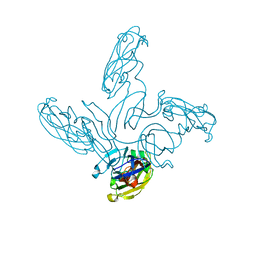 | | Crystal structure of the stem and receptor binding domain of the spike protein P1 from bacteriophage PM2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, SPIKE PROTEIN P1 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2VVF
 
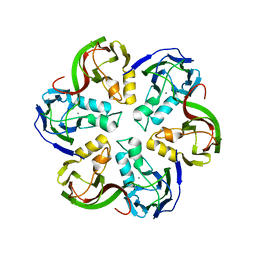 | | Crystal structure of the major capsid protein P2 from Bacteriophage PM2 | | Descriptor: | CALCIUM ION, MAJOR CAPSID PROTEIN P2 | | Authors: | Abrescia, N.G.A, Grimes, J.M, Kivela, H.K, Assenberg, R, Sutton, G.C, Butcher, S.J, Bamford, J.K.H, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2008-06-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insights Into Virus Evolution and Membrane Biogenesis from the Structure of the Marine Lipid-Containing Bacteriophage Pm2.
Mol.Cell, 31, 2008
|
|
2VVX
 
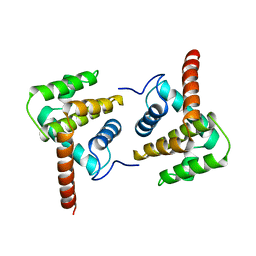 | | Structure of Vaccinia virus protein A52 | | Descriptor: | PROTEIN A52 | | Authors: | Graham, S.C, Bahar, M.W, Cooray, S, Chen, R.A.-J, Whalen, D.M, Abrescia, N.G.A, Alderton, D, Owens, R.J, Stuart, D.I, Smith, G.L, Grimes, J.M. | | Deposit date: | 2008-06-12 | | Release date: | 2008-08-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.746 Å) | | Cite: | Vaccinia Virus Proteins A52 and B14 Share a Bcl-2-Like Fold But Have Evolved to Inhibit NF-kappaB Rather Than Apoptosis
Plos Pathog., 4, 2008
|
|
2WUX
 
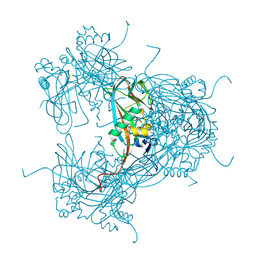 | | the crystal structure of recombinant baculovirus polyhedra | | Descriptor: | POLYHEDRIN | | Authors: | Ji, X, Sutton, G, Evans, G, Axford, D, Owen, R, Stuart, D.I. | | Deposit date: | 2009-10-10 | | Release date: | 2009-12-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | How Baculovirus Polyhedra Fit Square Pegs Into Round Holes to Robustly Package Viruses.
Embo J., 29, 2010
|
|
2WO2
 
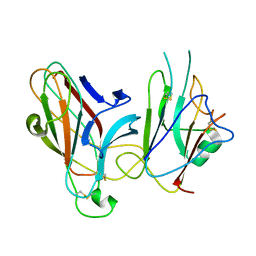 | | Crystal Structure of the EphA4-ephrinB2 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, EPHRIN TYPE-A RECEPTOR, EPHRIN-B2 | | Authors: | Bowden, T.A, Aricescu, A.R, Nettleship, J.E, Siebold, C, Rahman-Huq, N, Owens, R.J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 2009-07-21 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Plasticity of Eph-Receptor A4 Facilitates Cross-Class Ephrin Signalling
Structure, 17, 2009
|
|
2VHQ
 
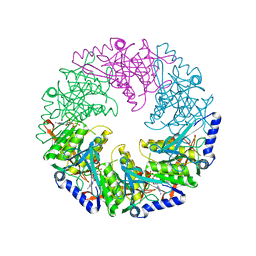 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ATP AND MG | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-22 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
2VHJ
 
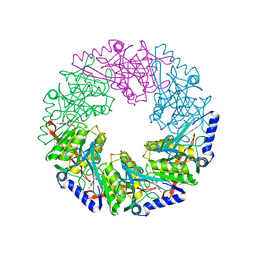 | | P4 PROTEIN FROM BACTERIOPHAGE PHI12 S252A mutant in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, NTPASE P4 | | Authors: | Kainov, D.E, Mancini, E.J, Telenius, J, Lisal, J, Grimes, J.M, Bamford, D.H, Stuart, D.I, Tuma, R. | | Deposit date: | 2007-11-21 | | Release date: | 2007-12-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Basis of Mechanochemical Coupling in a Hexameric Molecular Motor.
J.Biol.Chem., 283, 2008
|
|
