1T6U
 
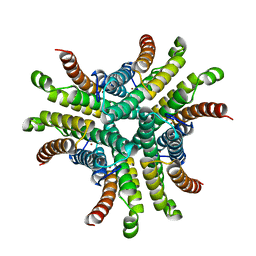 | | Nickel Superoxide Dismutase (NiSOD) Native 1.30 A Structure | | Descriptor: | NICKEL (II) ION, Superoxide dismutase [Ni] | | Authors: | Barondeau, D.P, Kassmann, C.J, Bruns, C.K, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2004-05-07 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Nickel superoxide dismutase structure and mechanism.
Biochemistry, 43, 2004
|
|
1TB0
 
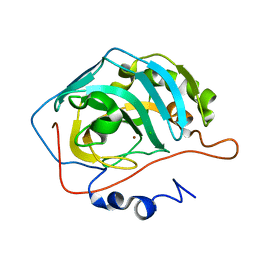 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-19 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of active-site histidine as a proton shuttle in catalysis by human carbonic anhydrase II.
Biochemistry, 44, 2005
|
|
4KUX
 
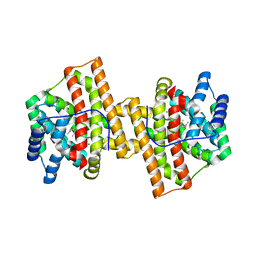 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | Aristolochene synthase, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Chen, M, Faraldos, J.A, Al-lami, N, Janvier, M, D'Antonio, E.L, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
1T6E
 
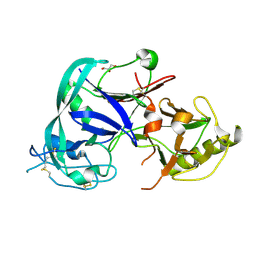 | | Crystal Structure of the Triticum aestivum xylanase inhibitor I | | Descriptor: | GLYCEROL, xylanase inhibitor | | Authors: | Sansen, S, De Ranter, C.J, Gebruers, K, Brijs, K, Courtin, C.M, Delcour, J.A, Rabijns, A. | | Deposit date: | 2004-05-06 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for inhibition of Aspergillus niger xylanase by triticum aestivum xylanase inhibitor-I
J.Biol.Chem., 279, 2004
|
|
1T9A
 
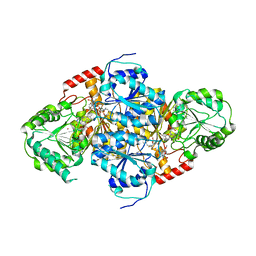 | | Crystal structure of yeast acetohydroxyacid synthase in complex with a sulfonylurea herbicide, tribenuron methyl | | Descriptor: | 2-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}PROPANE-1-THIOL, 5-{[ETHYL(METHYL)AMINO]METHYL}-2-METHYL-5,6-DIHYDROPYRIMIDIN-4-AMINE, Acetolactate synthase, ... | | Authors: | McCourt, J.A, Pang, S.S, Guddat, L.W, Duggleby, R.G. | | Deposit date: | 2004-05-16 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Elucidating the specificity of binding of sulfonylurea herbicides to acetohydroxyacid synthase.
Biochemistry, 44, 2005
|
|
4L0N
 
 | | Crystal structure of STK3 (MST2) SARAH domain | | Descriptor: | Serine/threonine-protein kinase 3 | | Authors: | Chaikuad, A, Krojer, T, Newman, J.A, Dixon-Clarke, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-05-31 | | Release date: | 2013-06-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of STK3 (MST2) SARAH domain
To be Published
|
|
1TE3
 
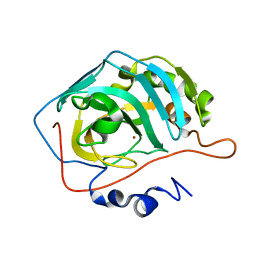 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | CHLORIDE ION, Carbonic anhydrase II, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-24 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1TEQ
 
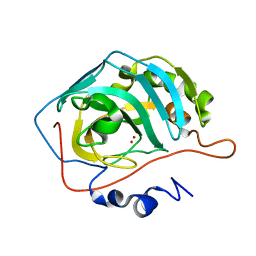 | | Effect of Shuttle Location and pH Environment on H+ Transfer in Human Carbonic Anhydrase II | | Descriptor: | Carbonic anhydrase II, HYDROXIDE ION, ZINC ION | | Authors: | Fisher, Z, Hernandez Prada, J.A, Tu, C.K, Duda, D, Yoshioka, C, An, H, Govindasamy, L, Silverman, D.N, McKenna, R. | | Deposit date: | 2004-05-25 | | Release date: | 2005-01-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Kinetic Characterization of Active-Site Histidine as a Proton Shuttle in Catalysis by Human Carbonic Anhydrase II
Biochemistry, 44, 2005
|
|
1VVJ
 
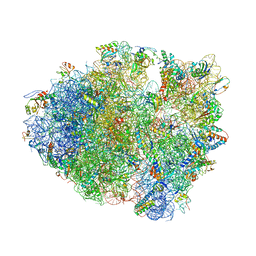 | | Crystal Structure of Frameshift Suppressor tRNA SufA6 bound to Codon CCC-G on the Ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Maehigashi, T, Dunkle, J.A, Dunham, C.M. | | Deposit date: | 2013-05-24 | | Release date: | 2014-08-06 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.440001 Å) | | Cite: | Structural insights into +1 frameshifting promoted by expanded or modification-deficient anticodon stem loops.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4KVI
 
 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (4aS,7S)-4a-methyl-7-(prop-1-en-2-yl)-2,3,4,4a,5,6,7,8-octahydroquinolin-1-ium | | Descriptor: | (4aS,7S)-4a-methyl-7-(prop-1-en-2-yl)-2,3,4,4a,5,6,7,8-octahydroquinolinium, Aristolochene synthase, MAGNESIUM ION, ... | | Authors: | Chen, M, Faraldos, J.A, Al-lami, N, Janvier, M, D'Antonio, E.L, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-22 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4KZD
 
 | | Crystal structure of an RNA aptamer in complex with fluorophore and Fab | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, BL3-6 Fab antibody, heavy chain, ... | | Authors: | Huang, H, Suslov, N.B, Li, N, Koldobskaya, Y, Rice, P.A, Piccirilli, J.A. | | Deposit date: | 2013-05-29 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.186 Å) | | Cite: | A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore.
Nat.Chem.Biol., 10, 2014
|
|
4L05
 
 | | Cu/Zn superoxide dismutase from Brucella abortus | | Descriptor: | COPPER (I) ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Shin, D.S, Didonato, M, Pratt, A.J, Bruns, C.K, Cabelli, D.E, Kroll, J.S, Belzer, C.A, Tabatabai, L.B, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2013-05-30 | | Release date: | 2013-10-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.098 Å) | | Cite: | Structural, Functional, and Immunogenic Insights on Cu,Zn Superoxide Dismutase Pathogenic Virulence Factors from Neisseria meningitidis and Brucella abortus.
J.Bacteriol., 197, 2015
|
|
1SZU
 
 | | The structure of gamma-aminobutyrate aminotransferase mutant: V241A | | Descriptor: | 1,2-ETHANEDIOL, 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-aminobutyrate aminotransferase, ... | | Authors: | Liu, W, Peterson, P.E, Langston, J.A, Jin, X, Zhou, X, Fisher, A.J, Toney, M.D. | | Deposit date: | 2004-04-06 | | Release date: | 2005-03-01 | | Last modified: | 2021-10-27 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Kinetic and Crystallographic Analysis of Active Site Mutants of Escherichia coligamma-Aminobutyrate Aminotransferase.
Biochemistry, 44, 2005
|
|
4LC8
 
 | | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 1,2-ETHANEDIOL, 6-HYDROXYURIDINE-5'-PHOSPHATE, CHLORIDE ION, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-06-21 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.318 Å) | | Cite: | Crystal structure of the mutant H128N of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP
To be Published
|
|
4KVW
 
 | | Crystal structure of Aspergillus terreus aristolochene synthase complexed with (3R,6R,9aR)-6,9a-dimethyl-3-(prop-1-en-2-yl)decahydroquinolizin-5-ium | | Descriptor: | (3R,5S,6R,9aR)-6,9a-dimethyl-3-(prop-1-en-2-yl)octahydro-2H-quinolizinium, Aristolochene synthase, GLYCEROL, ... | | Authors: | Chen, M, Al-lami, N, Janvier, M, D'Antonio, E.L, Faraldos, J.A, Cane, D.E, Allemann, R.K, Christianson, D.W. | | Deposit date: | 2013-05-23 | | Release date: | 2013-08-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Mechanistic insights from the binding of substrate and carbocation intermediate analogues to aristolochene synthase.
Biochemistry, 52, 2013
|
|
4KWS
 
 | | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg and glycerol | | Descriptor: | CHLORIDE ION, D-mannonate dehydratase, GLYCEROL, ... | | Authors: | Fedorov, A.A, Fedorov, E.V, Wichelecki, D, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2013-05-24 | | Release date: | 2014-05-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.642 Å) | | Cite: | Crystal structure of d-mannonate dehydratase from chromohalobacter salexigens complexed with Mg and glycerol
To be Published
|
|
1W3A
 
 | | Three dimensional structure of a novel pore-forming lectin from the mushroom Laetiporus sulphureus | | Descriptor: | GLYCEROL, HEMOLYTIC LECTIN LSLA, beta-D-galactopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Mancheno, J.M, Tateno, H, Goldstein, I.J, Martinez-Ripoll, M, Hermoso, J.A. | | Deposit date: | 2004-07-14 | | Release date: | 2005-02-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural Analysis of the Laetiporus Sulphureus Hemolytic Pore-Forming Lectin in Complex with Sugars
J.Biol.Chem., 280, 2005
|
|
1W3F
 
 | | Crystal structure of the hemolytic lectin from the mushroom Laetiporus sulphureus complexed with N-acetyllactosamine in the gamma motif | | Descriptor: | GLYCEROL, HEMOLYTIC LECTIN FROM LAETIPORUS SULPHUREUS, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Mancheno, J.M, Tateno, H, Goldstein, I.J, Martinez-Ripoll, M, Hermoso, J.A. | | Deposit date: | 2004-07-15 | | Release date: | 2005-02-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural Analysis of the Laetiporus Sulphureus Hemolytic Pore-Forming Lectin in Complex with Sugars
J.Biol.Chem., 280, 2005
|
|
3QMR
 
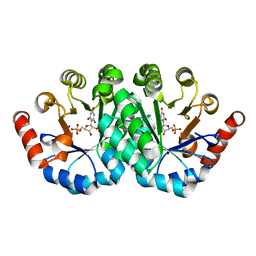 | | Crystal structure of the mutant R160A,V182A of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3213 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
7JRP
 
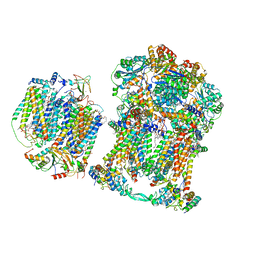 | | Plant Mitochondrial complex SC III2+IV from Vigna radiata | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, Alpha-MPP, ... | | Authors: | Maldonado, M, Letts, J.A. | | Deposit date: | 2020-08-12 | | Release date: | 2021-01-20 | | Last modified: | 2021-02-03 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structures of respiratory complex III 2 , complex IV, and supercomplex III 2 -IV from vascular plants.
Elife, 10, 2021
|
|
7KDF
 
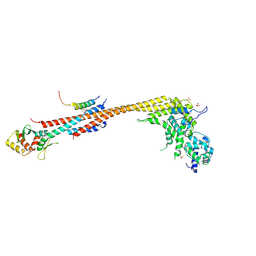 | | Structure of Stu2 Bound to dwarf Ndc80c | | Descriptor: | NDC80 isoform 1,NDC80 isoform 1, NUF2 isoform 1,NUF2 isoform 1, SPC25 isoform 1,SPC25 isoform 1, ... | | Authors: | Zahm, J.A, Stewart, M.G, Miller, M.P, Harrison, S.C. | | Deposit date: | 2020-10-08 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structural basis of Stu2 recruitment to yeast kinetochores.
Elife, 10, 2021
|
|
3QKR
 
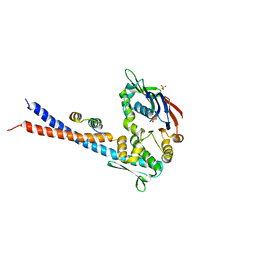 | | Mre11 Rad50 binding domain bound to Rad50 | | Descriptor: | DNA double-strand break repair protein mre11, DNA double-strand break repair rad50 ATPase, PHOSPHATE ION | | Authors: | Williams, G.J, Williams, R.S, Arvai, A, Moncalian, G, Tainer, J.A. | | Deposit date: | 2011-02-01 | | Release date: | 2011-03-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair.
Nat.Struct.Mol.Biol., 18, 2011
|
|
3QMT
 
 | | Crystal structure of the mutant V182A,Y206F of orotidine 5'-monophosphate decarboxylase from Methanobacterium thermoautotrophicum complexed with the inhibitor BMP | | Descriptor: | 6-HYDROXYURIDINE-5'-PHOSPHATE, GLYCEROL, Orotidine 5'-phosphate decarboxylase | | Authors: | Fedorov, A.A, Fedorov, E.V, Desai, B, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2011-02-05 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.3202 Å) | | Cite: | Conformational changes in orotidine 5'-monophosphate decarboxylase: a structure-based explanation for how the 5'-phosphate group activates the enzyme.
Biochemistry, 51, 2012
|
|
3QN1
 
 | | Crystal structure of the PYR1 Abscisic Acid receptor in complex with the HAB1 type 2C phosphatase catalytic domain | | Descriptor: | (2Z,4E)-5-[(1S)-1-hydroxy-2,6,6-trimethyl-4-oxocyclohex-2-en-1-yl]-3-methylpenta-2,4-dienoic acid, Abscisic acid receptor PYR1, MANGANESE (II) ION, ... | | Authors: | Betz, K, Dupeux, F, Santiago, J, Marquez, J.A. | | Deposit date: | 2011-02-07 | | Release date: | 2011-03-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Modulation of Abscisic Acid Signaling in Vivo by an Engineered Receptor-Insensitive Protein Phosphatase Type 2C Allele.
Plant Physiol., 156, 2011
|
|
7JX2
 
 | |
