5BP0
 
 | | X-ray crystal structure of Lymnaea stagnalis acetylcholine binding protein (Ls-AChBP) in complex with 5-Fluoronicotine (TI-4650) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 5-fluoronicotine, Acetylcholine-binding protein, ... | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-05-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
7WKJ
 
 | | A COVID-19 T-cell response detection method based on a newly identified human CD8+ T cell epitope from SARS-CoV-2-Hubei Province, 2021. | | Descriptor: | Beta-2-microglobulin, LYS-THR-PHE-PRO-PRO-THR-GLU-PRO-LYS, MHC class I antigen | | Authors: | Zhang, J, Lu, D, Li, M, Liu, M.S, Yao, S.J, Zhan, J.B, Liu, J, Gao, G.F. | | Deposit date: | 2022-01-10 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A COVID-19 T-Cell Response Detection Method Based on a Newly Identified Human CD8 + T Cell Epitope from SARS-CoV-2 - Hubei Province, China, 2021.
China CDC Wkly, 4, 2022
|
|
5BRX
 
 | | X-ray crystal structure of Aplysia californica (Ac-AChBP) in complex with 2-pyridyl azatricyclo[3.3.1.13,7]decane; 2-pyridylazaadamantane; 2-Aza (TI-8480) | | Descriptor: | (2R,3S,5R,7S)-2-(pyridin-3-yl)-1-azatricyclo[3.3.1.1~3,7~]decane, SULFATE ION, Soluble acetylcholine receptor, ... | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-06-01 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
8ILJ
 
 | |
6DGL
 
 | |
6K3E
 
 | | LSD1/Co-Rest structure with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-PCPA derivative, ... | | Authors: | Wang, J. | | Deposit date: | 2019-05-17 | | Release date: | 2020-05-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | LSD1/Co-Rest structure with an inhibitor
To Be Published
|
|
5ZY9
 
 | | Structural basis for a tRNA synthetase | | Descriptor: | (1R,2R)-2-[(2S,4E,6E,8R,9S,11R,13S,15S,16S)-7-cyano-8,16-dihydroxy-9,11,13,15-tetramethyl-18-oxooxacyclooctadeca-4,6-dien-2-yl]cyclopentanecarboxylic acid, Threonyl-tRNA synthase, ZINC ION | | Authors: | Zhang, J. | | Deposit date: | 2018-05-23 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for inhibition of
translational functions on a tRNA synthetase
To Be Published
|
|
6DGR
 
 | |
4IVN
 
 | | The Vibrio vulnificus NanR protein complexed with ManNAc-6P | | Descriptor: | 2-acetamido-2-deoxy-6-O-phosphono-alpha-D-mannopyranose, Transcriptional regulator | | Authors: | Hwang, J, Kim, M.H. | | Deposit date: | 2013-01-23 | | Release date: | 2013-07-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into the regulation of sialic acid catabolism by the Vibrio vulnificus transcriptional repressor NanR
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7X89
 
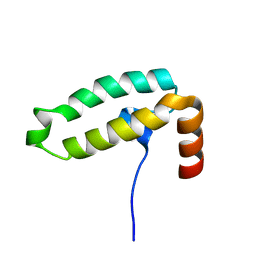 | | Tid1 | | Descriptor: | DnaJ homolog subfamily A member 3, mitochondrial | | Authors: | Jang, J, Lee, S.H, Kang, D.H, Sim, D.W, Jo, K.S, Ryu, H, Kim, E.H, Ryu, K.S, Lee, J.H, Kim, J.H, Won, H.S. | | Deposit date: | 2022-03-11 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural studies on the J-domain and GF-motif of the mitochondrial Hsp40, Tid1
To Be Published
|
|
8JUO
 
 | |
6LVO
 
 | |
8JUS
 
 | |
6LP3
 
 | | Structural basis and functional analysis epo1-bem3p complex for bud growth | | Descriptor: | GTPase-activating protein BEM3, Uncharacterized protein YMR124W | | Authors: | Wang, J, Li, L, Ming, Z.H, Wu, L.J, Yan, L.M. | | Deposit date: | 2020-01-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.547 Å) | | Cite: | Structural basis and functional analysis epo1-bem3p complex for bud growth
To Be Published
|
|
6DGP
 
 | |
6DGQ
 
 | | Crystal Structure of Human PPARgamma Ligand Binding Domain in Complex with CAY10506 | | Descriptor: | N-(2-{4-[(2,4-dioxo-3,4-dihydro-2H-1lambda~4~,3-thiazol-5-yl)methyl]phenoxy}ethyl)-5-[(3R)-1,2-dithiolan-3-yl]pentanamide, Peroxisome proliferator-activated receptor gamma | | Authors: | Shang, J, Kojetin, D.J. | | Deposit date: | 2018-05-17 | | Release date: | 2019-05-22 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Quantitative structural assessment of graded receptor agonism.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6LP4
 
 | |
3J9C
 
 | | CryoEM single particle reconstruction of anthrax toxin protective antigen pore at 2.9 Angstrom resolution | | Descriptor: | CALCIUM ION, Protective antigen PA-63 | | Authors: | Jiang, J, Pentelute, B.L, Collier, R.J, Zhou, Z.H. | | Deposit date: | 2014-12-25 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Atomic structure of anthrax protective antigen pore elucidates toxin translocation.
Nature, 521, 2015
|
|
2LR7
 
 | | Cathelicidin-PY | | Descriptor: | Cathelicidin-PY | | Authors: | Yang, J. | | Deposit date: | 2012-03-27 | | Release date: | 2013-03-27 | | Method: | SOLUTION NMR | | Cite: | Structure of Cathelicidin-PY
To be Published
|
|
6BIE
 
 | |
6CWE
 
 | | Structure of alpha-GSA[8,6P] bound by CD1d and in complex with the Va14Vb8.2 TCR | | Descriptor: | (5R,6S,7S)-5,6-dihydroxy-7-(octanoylamino)-N-(6-phenylhexyl)-8-{[(2S,3R,4S,5R,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl]oxy}octanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wang, J, Zajonc, D. | | Deposit date: | 2018-03-30 | | Release date: | 2019-04-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Glycolipid recognition by iNKT cells
To Be Published
|
|
5BW2
 
 | | X-ray crystal structure of Aplysia californica acetylcholine binding protein (Ac-AChBP) Y55W in complex with 2-Pyridin-3-yl-1-aza-bicyclo[2.2.2]octane; 2-(3-pyridyl)quinuclidine; 2-PQ (TI-4699) | | Descriptor: | (2R)-2-(pyridin-3-yl)-1-azabicyclo[2.2.2]octane, Soluble acetylcholine receptor | | Authors: | Bobango, J, Sankaran, B, Park, J.F, Wu, J, Talley, T.T. | | Deposit date: | 2015-06-05 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Comparisons of Binding Affinities for Neuronal Nicotinic Receptors (NNRs) and AChBPs
To Be Published
|
|
1XX5
 
 | | Crystal Structure of Natrin from Naja atra snake venom | | Descriptor: | ETHANOL, Natrin 1 | | Authors: | Wang, J, Shen, B, Lou, X.H, Guo, M, Teng, M.K, Niu, L.W. | | Deposit date: | 2004-11-04 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Blocking effect and crystal structure of natrin toxin, a cysteine-rich secretory protein from Naja atra venom that targets the BKCa channel
Biochemistry, 44, 2005
|
|
2NWM
 
 | |
2KZL
 
 | |
