3A4N
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) kinase | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, L-seryl-tRNA(Sec) kinase, ... | | Authors: | Araiso, Y, Ishitani, R, Soll, D, Nureki, O. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a tRNA-dependent kinase essential for selenocysteine decoding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3AL6
 
 | | Crystal structure of Human TYW5 | | Descriptor: | 2-OXOGLUTARIC ACID, JmjC domain-containing protein C2orf60, NICKEL (II) ION | | Authors: | Kato, M, Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2010-07-26 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification.
Nucleic Acids Res., 39, 2011
|
|
3A4L
 
 | | Crystal structure of archaeal O-phosphoseryl-tRNA(Sec) kinase | | Descriptor: | 1,2-ETHANEDIOL, IODIDE ION, L-seryl-tRNA(Sec) kinase, ... | | Authors: | Araiso, Y, Ishitani, R, Soll, D, Nureki, O. | | Deposit date: | 2009-07-10 | | Release date: | 2009-10-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of a tRNA-dependent kinase essential for selenocysteine decoding
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2ZNI
 
 | | Crystal structure of Pyrrolysyl-tRNA synthetase-tRNA(Pyl) complex from Desulfitobacterium hafniense | | Descriptor: | CALCIUM ION, Pyrrolysyl-tRNA synthetase, bacterial tRNA | | Authors: | Nozawa, K, Araiso, Y, Soll, D, Ishitani, R, Nureki, O. | | Deposit date: | 2008-04-25 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality
Nature, 457, 2009
|
|
2ZNJ
 
 | | Crystal structure of Pyrrolysyl-tRNA synthetase from Desulfitobacterium hafniense | | Descriptor: | Putative uncharacterized protein | | Authors: | Nozawa, K, Araiso, Y, Soll, D, Ishitani, R, Nureki, O. | | Deposit date: | 2008-04-25 | | Release date: | 2008-12-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pyrrolysyl-tRNA synthetase-tRNA(Pyl) structure reveals the molecular basis of orthogonality
Nature, 457, 2009
|
|
2ZWA
 
 | | Crystal structure of tRNA wybutosine synthesizing enzyme TYW4 | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, Leucine carboxyl methyltransferase 2, ... | | Authors: | Suzuki, Y, Noma, A, Suzuki, T, Ishitani, R, Nureki, O. | | Deposit date: | 2008-12-01 | | Release date: | 2009-06-02 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of tRNA modification with CO2 fixation and methylation by wybutosine synthesizing enzyme TYW4.
Nucleic Acids Res., 37, 2009
|
|
3A1T
 
 | |
2ZZE
 
 | | Crystal structure of alanyl-tRNA synthetase without oligomerization domain in lysine-methylated form | | Descriptor: | Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3AY5
 
 | |
2YVX
 
 | | Crystal structure of magnesium transporter MgtE | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
3AL5
 
 | | Crystal structure of Human TYW5 | | Descriptor: | JmjC domain-containing protein C2orf60, MAGNESIUM ION | | Authors: | Kato, M, Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2010-07-26 | | Release date: | 2010-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.503 Å) | | Cite: | Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification.
Nucleic Acids Res., 39, 2011
|
|
6LEH
 
 | | Crystal structure of Autotaxin in complex with an inhibitor | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Nishimasu, H, Osamu, N. | | Deposit date: | 2019-11-25 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of PotentIn VivoAutotaxin Inhibitors that Bind to Both Hydrophobic Pockets and Channels in the Catalytic Domain.
J.Med.Chem., 63, 2020
|
|
6LBH
 
 | |
5Y5F
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by low-dose (0.72 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5H
 
 | | SF-ROX structure of cytochrome P450nor (NO-bound state) determined at SACLA | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Yamagiwa, R, Yamashita, K, Hirata, K, Ueno, G, Kimura, T, Hisano, T, Muramoto, K, Sawai, H, Takeda, H, Yamashita, A, Murakami, H, Owada, S, Tono, K, Yabashi, M, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
5Y5G
 
 | | Structure of cytochrome P450nor in NO-bound state: damaged by high-dose (5.7 MGy) X-ray | | Descriptor: | GLYCEROL, NADP nitrous oxide-forming nitric oxide reductase, NITRIC OXIDE, ... | | Authors: | Tosha, T, Nomura, T, Nishida, T, Ueno, G, Murakami, H, Yamashita, K, Hirata, K, Yamamoto, M, Ago, H, Sugimoto, H, Shiro, Y, Kubo, M. | | Deposit date: | 2017-08-09 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Capturing an initial intermediate during the P450nor enzymatic reaction using time-resolved XFEL crystallography and caged-substrate.
Nat Commun, 8, 2017
|
|
7Q7P
 
 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | Authors: | Baath, P, Banacore, A, Neutze, R. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q7Q
 
 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | Authors: | Baath, P, Banacore, A, Neutze, R. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
8AJZ
 
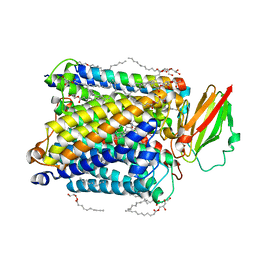 | | Serial femtosecond crystallography structure of CO bound ba3- type cytochrome c oxidase at 2 milliseconds after irradiation by a 532 nm laser | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CARBON MONOXIDE, COPPER (II) ION, ... | | Authors: | Safari, C, Ghosh, S, Andersson, R, Johannesson, J, Donoso, A.V, Bath, P, Bosman, R, Dahl, P, Nango, E, Tanaka, R, Zoric, D, Svensson, E, Nakane, T, Iwata, S, Neutze, R, Branden, G. | | Deposit date: | 2022-07-29 | | Release date: | 2023-08-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Time-resolved serial crystallography to track the dynamics of carbon monoxide in the active site of cytochrome c oxidase.
Sci Adv, 9, 2023
|
|
5JOO
 
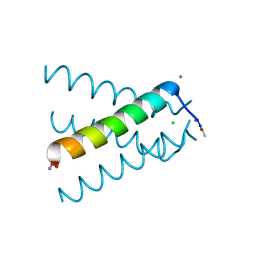 | | XFEL structure of influenza A M2 wild type TM domain at low pH in the lipidic cubic phase at room temperature | | Descriptor: | CALCIUM ION, CHLORIDE ION, Matrix protein 2 | | Authors: | Thomaston, J.L, Woldeyes, R.A, Fraser, J.S, DeGrado, W.F. | | Deposit date: | 2016-05-02 | | Release date: | 2017-08-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.413 Å) | | Cite: | XFEL structures of the influenza M2 proton channel: Room temperature water networks and insights into proton conduction.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5KXU
 
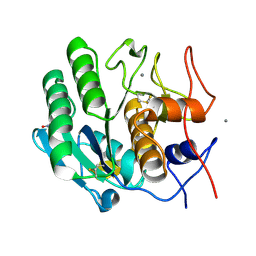 | | Structure Proteinase K determined by SACLA | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
5KXV
 
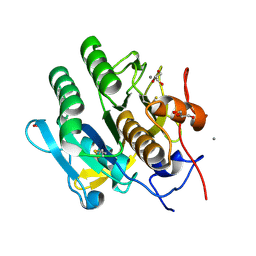 | | Structure Proteinase K at 0.98 Angstroms | | Descriptor: | CALCIUM ION, GLYCEROL, NITRATE ION, ... | | Authors: | Masuda, T, Suzuki, M, Inoue, S, Numata, K, Sugahara, M. | | Deposit date: | 2016-07-20 | | Release date: | 2017-06-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Atomic resolution structure of serine protease proteinase K at ambient temperature.
Sci Rep, 7, 2017
|
|
3H39
 
 | | The complex structure of CCA-adding enzyme with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, TRNA nucleotidyl transferase-related protein | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.854 Å) | | Cite: | Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme
Embo J., 28, 2009
|
|
1WQ3
 
 | | Escherichia coli tyrosyl-tRNA synthetase mutant complexed with 3-iodo-L-tyrosine | | Descriptor: | 3-IODO-TYROSINE, Tyrosyl-tRNA synthetase | | Authors: | Kobayashi, T, Sakamoto, K, Nureki, O, Takimura, T, Kamata, K, Sekine, R, Nishimura, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-20 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of nonnatural amino acid recognition by an engineered aminoacyl-tRNA synthetase for genetic code expansion
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
3H38
 
 | | The structure of CCA-adding enzyme apo form II | | Descriptor: | TRNA nucleotidyl transferase-related protein | | Authors: | Toh, Y, Tomita, K. | | Deposit date: | 2009-04-16 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Mechanism for the definition of elongation and termination by the class II CCA-adding enzyme
Embo J., 28, 2009
|
|
