3HOS
 
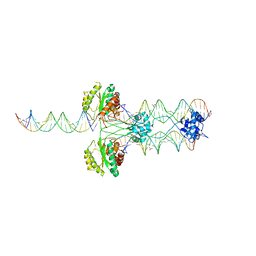 | |
3HVD
 
 | | The Protective Antigen Component of Anthrax Toxin Forms Functional Octameric Complexes | | Descriptor: | CALCIUM ION, Protective antigen | | Authors: | Kintzer, A.F, Dong, K.C, Berger, J.M, Krantz, B.A. | | Deposit date: | 2009-06-15 | | Release date: | 2009-08-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.209 Å) | | Cite: | The protective antigen component of anthrax toxin forms functional octameric complexes.
J.Mol.Biol., 392, 2009
|
|
3HY9
 
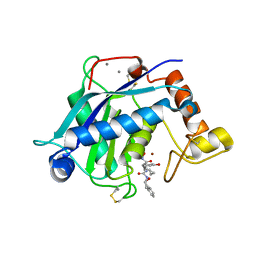 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with an Amino-2-indanol compound | | Descriptor: | (3R)-N~2~-(cyclopropylmethyl)-N~1~-hydroxy-3-(3-hydroxybenzyl)-N~4~-[(1S,2R)-2-hydroxy-2,3-dihydro-1H-inden-1-yl]-L-aspartamide, CALCIUM ION, Catalytic Domain of ADAMTS-5, ... | | Authors: | Shieh, H.-S, Williams, J.M, Caspers, N, Mathis, K.J, Tortorella, M.D, Tomasselli, A. | | Deposit date: | 2009-06-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural and inhibition analysis reveals the mechanism of selectivity of a series of aggrecanase inhibitors
J.Biol.Chem., 284, 2009
|
|
3I4K
 
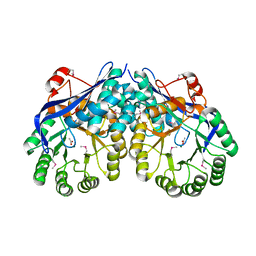 | | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum | | Descriptor: | ACETIC ACID, MAGNESIUM ION, Muconate lactonizing enzyme | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Muconate lactonizing enzyme from Corynebacterium glutamicum
To be Published
|
|
3HU5
 
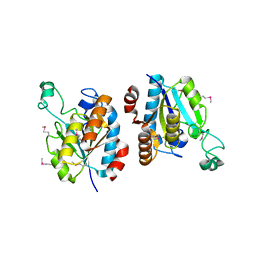 | | CRYSTAL STRUCTURE OF isochorismatase family protein from Desulfovibrio vulgaris subsp. vulgaris str. Hildenborough | | Descriptor: | Isochorismatase family protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-06-12 | | Release date: | 2009-06-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CRYSTAL STRUCTURE OF isochorismatase family protein from Desulfovibrio vulgaris subsp.
vulgaris str. Hildenborough
To be Published
|
|
3I4J
 
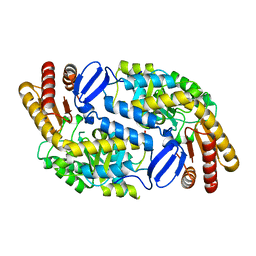 | | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans | | Descriptor: | Aminotransferase, class III, SULFATE ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Aminotransferase, class III from Deinococcus radiodurans
To be Published
|
|
3I9V
 
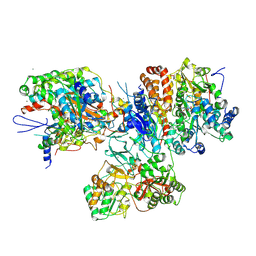 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, oxidized, 2 mol/ASU | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-13 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
3IRV
 
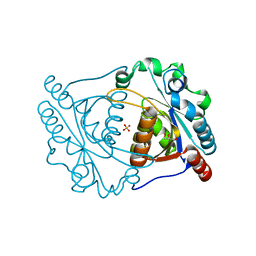 | | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae pv. phaseolicola 1448A | | Descriptor: | CYSTEINE HYDROLASE, GLYCEROL, PHOSPHATE ION | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF CYSTEINE HYDROLASE PSPPH_2384 FROM Pseudomonas syringae
To be Published
|
|
3IJD
 
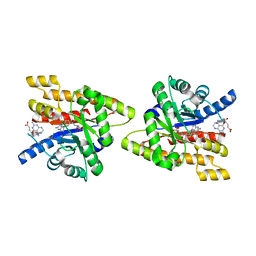 | | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid | | Descriptor: | 5-METHYL-5,6,7,8-TETRAHYDROFOLIC ACID, uncharacterized protein | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-04 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Uncharacterized protein Cthe_2304 from Clostridium thermocellum binds two copies of 5-methyl-5,6,7,8-tetrahydrofolic acid
To be Published
|
|
3ISA
 
 | | CRYSTAL STRUCTURE OF putative enoyl-CoA hydratase/isomerase FROM Bordetella parapertussis | | Descriptor: | CHLORIDE ION, GLYCEROL, Putative enoyl-CoA hydratase/isomerase | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-25 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF enoyl-CoA hydratase FROM Bordetella parapertussis
To be Published
|
|
3I13
 
 | |
3I45
 
 | | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum Atcc 11170 | | Descriptor: | NICOTINIC ACID, Twin-arginine translocation pathway signal protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | CRYSTAL STRUCTURE OF putative twin-arginine translocation pathway signal protein from Rhodospirillum rubrum
Atcc 11170
To be Published
|
|
3I6E
 
 | | CRYSTAL STRUCTURE OF MUCONATE LACTONIZING ENZYME FROM Ruegeria pomeroyi. | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase I, SODIUM ION | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of muconate lactonizing enzyme from Ruegeria pomeroyi.
To be Published
|
|
3FBE
 
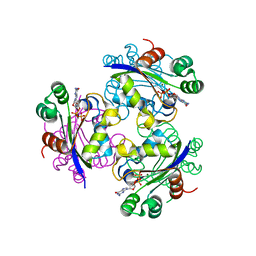 | | Crystal structure of the Mimivirus NDK N62L-R107G double mutant complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Nucleoside diphosphate kinase | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3F5Q
 
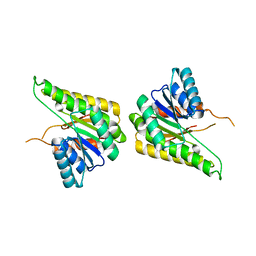 | | CRYSTAL STRUCTURE OF putative short chain dehydrogenase from Escherichia coli CFT073 | | Descriptor: | dehydrogenase | | Authors: | Malashkevich, V.N, Toro, R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-04 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of an uncharacterized protein
to be published
|
|
3FBC
 
 | | Crystal structure of the Mimivirus NDK N62L-R107G double mutant complexed with dTDP | | Descriptor: | MAGNESIUM ION, Nucleoside diphosphate kinase, PHOSPHATE ION, ... | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-19 | | Release date: | 2009-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3FC9
 
 | | Crystal structure of the Mimivirus NDK +Kpn-N62L double mutant complexed with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, CYTIDINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Jeudy, S, Lartigue, A, Claverie, J.M, Abergel, C. | | Deposit date: | 2008-11-21 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dissecting the unique nucleotide specificity of mimivirus nucleoside diphosphate kinase.
J.Virol., 83, 2009
|
|
3FDU
 
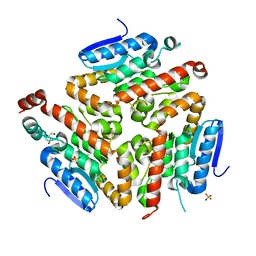 | | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii | | Descriptor: | GLYCEROL, Putative enoyl-CoA hydratase/isomerase, SULFATE ION | | Authors: | Bonanno, J.B, Dickey, M, Bain, K.T, Tang, B.K, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-26 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a putative enoyl-CoA hydratase/isomerase from Acinetobacter baumannii
To be Published
|
|
3FEQ
 
 | | Crystal structure of uncharacterized protein eah89906 | | Descriptor: | PUTATIVE AMIDOHYDROLASE, ZINC ION | | Authors: | Patskovsky, Y, Bonanno, J, Romero, R, Freeman, J, Lau, C, Smith, D, Bain, K, Wasserman, S.R, Raushel, F, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-30 | | Release date: | 2008-12-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.63 Å) | | Cite: | Functional identification and structure determination of two novel prolidases from cog1228 in the amidohydrolase superfamily .
Biochemistry, 49, 2010
|
|
3IAS
 
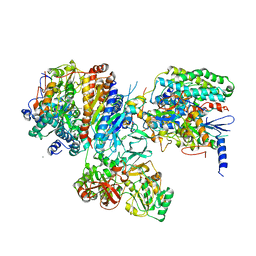 | | Crystal structure of the hydrophilic domain of respiratory complex I from Thermus thermophilus, oxidized, 4 mol/ASU, re-refined to 3.15 angstrom resolution | | Descriptor: | CALCIUM ION, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Sazanov, L.A, Berrisford, J.M. | | Deposit date: | 2009-07-14 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for the mechanism of respiratory complex I
J.Biol.Chem., 284, 2009
|
|
3IWA
 
 | | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris | | Descriptor: | CALCIUM ION, FAD-dependent pyridine nucleotide-disulphide oxidoreductase | | Authors: | Bonanno, J.B, Freeman, J, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-09-02 | | Release date: | 2009-10-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a FAD-dependent pyridine nucleotide-disulphide oxidoreductase from Desulfovibrio vulgaris
To be Published
|
|
3IAN
 
 | | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis | | Descriptor: | 1,2-ETHANEDIOL, Chitinase, SODIUM ION | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Miller, S, Ozyurt, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-14 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a chitinase from Lactococcus lactis subsp. lactis
To be Published
|
|
3IBM
 
 | | CRYSTAL STRUCTURE OF cupin 2 domain-containing protein Hhal_0468 FROM Halorhodospira halophila | | Descriptor: | Cupin 2, conserved barrel domain protein, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Freeman, J, Miller, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-16 | | Release date: | 2009-07-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF cupin 2 domain-containing PROTEIN Hhal_0468 FROM Halorhodospira halophila
To be Published
|
|
3ICJ
 
 | | Crystal structure of an uncharacterized metal-dependent hydrolase from pyrococcus furiosus | | Descriptor: | ZINC ION, uncharacterized metal-dependent hydrolase | | Authors: | Bonanno, J.B, Patskovsky, Y, Freeman, J, Bain, K.T, Hu, S, Ozyurt, S, Wasserman, S, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-07-17 | | Release date: | 2009-07-28 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of an Uncharacterized Metal-Dependent Hydrolase from Pyrococcus Furiosus
To be Published
|
|
3IRS
 
 | | CRYSTAL STRUCTURE OF UNCHARACTERIZED TIM-BARREL PROTEIN BB4693 FROM Bordetella bronchiseptica | | Descriptor: | GLYCEROL, SODIUM ION, SULFATE ION, ... | | Authors: | Patskovsky, Y, Malashkevich, V, Toro, R, Foti, R, Dickey, M, Do, J, Sauder, J.M, Raushel, F.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-08-24 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | CRYSTAL STRUCTURE OF UNCHARACTERIZED HYDROLASE FROM Bordetella bronchiseptica
To be Published
|
|
