2V5U
 
 | | I92A FLAVODOXIN FROM ANABAENA | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN | | Authors: | Martinez-Julvez, M, Herguedas, B, Frago, S, Serrano, A, Molina, R, Hamiaux, C, Schierbeek, B, Medina, M, Hermoso, J.A. | | Deposit date: | 2007-07-10 | | Release date: | 2007-10-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Tuning of the Fmn Binding and Oxido-Reduction Properties by Neighboring Side Chains in Anabaena Flavodoxin.
Arch.Biochem.Biophys., 467, 2007
|
|
1QUE
 
 | | X-RAY STRUCTURE OF THE FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 1.8 ANGSTROMS | | Descriptor: | FERREDOXIN--NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-07-06 | | Release date: | 1997-05-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
1QUF
 
 | | X-RAY STRUCTURE OF A COMPLEX NADP+-FERREDOXIN:NADP+ REDUCTASE FROM THE CYANOBACTERIUM ANABAENA PCC 7119 AT 2.25 ANGSTROMS | | Descriptor: | FERREDOXIN-NADP+ REDUCTASE, FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Serre, L, Frey, M, Vellieux, F.M.D. | | Deposit date: | 1996-09-07 | | Release date: | 1997-09-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | X-ray structure of the ferredoxin:NADP+ reductase from the cyanobacterium Anabaena PCC 7119 at 1.8 A resolution, and crystallographic studies of NADP+ binding at 2.25 A resolution.
J.Mol.Biol., 263, 1996
|
|
3ESX
 
 | | E16KE61KD126KD150K Flavodoxin from Anabaena | | Descriptor: | CALCIUM ION, CHLORIDE ION, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Herguedas, B, Hermoso, J.A, Martinez-Julvez, M, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
4ILC
 
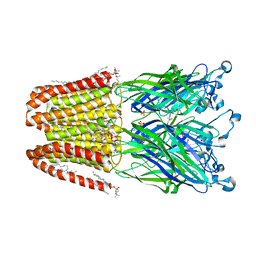 | | The GLIC pentameric ligand-gated ion channel in complex with sulfates | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
3ESY
 
 | | E16KE61K Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavodoxin, GLYCEROL | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J.A, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
4IO8
 
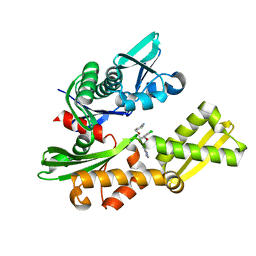 | | Crystal structure of human HSP70 complexed with 4-{(2R,3S,4R)-5-[(R)-6-Amino-8-(3,4-dichloro-benzylamino)-purin-9-yl]-3,4-dihydroxy-tetrahydro-furan-2-ylmethoxymethyl}-benzonitrile | | Descriptor: | 4-[[(2R,3S,4R,5R)-5-[6-amino-8-[(3,4-dichlorophenyl)methylamino]purin-9-yl]-3,4-dihydroxy-oxolan-2-yl]methoxymethyl]benzonitrile, Heat shock 70kDa protein 1A variant | | Authors: | Musil, D, Scholz, S. | | Deposit date: | 2013-01-07 | | Release date: | 2013-12-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Functional analysis of hsp70 inhibitors.
Plos One, 8, 2013
|
|
4IL4
 
 | | The pentameric ligand-gated ion channel GLIC in complex with Se-DDM | | Descriptor: | ACETATE ION, CHLORIDE ION, Proton-gated ion channel, ... | | Authors: | Sauguet, L, Malherbe, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4IL9
 
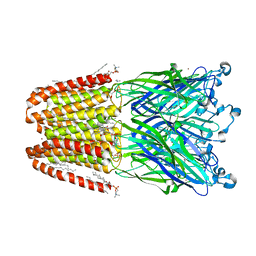 | | The pentameric ligand-gated ion channel GLIC A237F in complex with bromide | | Descriptor: | ACETATE ION, BROMIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4ILA
 
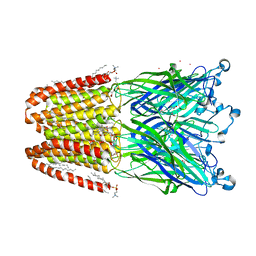 | | The pentameric ligand-gated ion channel GLIC A237F in complex with Cesium | | Descriptor: | ACETATE ION, CESIUM ION, CHLORIDE ION, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
3F8G
 
 | | The X-ray structure of a dimeric variant of human pancreatic ribonuclease with high cytotoxic and antitumor activities | | Descriptor: | Ribonuclease pancreatic, SULFATE ION | | Authors: | Merlino, A, Avella, G, Mazzarella, L, Sica, F. | | Deposit date: | 2008-11-12 | | Release date: | 2009-02-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural features for the mechanism of antitumor action of a dimeric human pancreatic ribonuclease variant.
Protein Sci., 18, 2009
|
|
5U2M
 
 | | Crystal structure of human NAMPT with A-1293201 | | Descriptor: | N-[4-({[(3S)-oxolan-3-yl]methyl}carbamoyl)phenyl]-1,3-dihydro-2H-isoindole-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
5U2N
 
 | | Crystal structure of human NAMPT with A-1326133 | | Descriptor: | N-{4-[1-(2-methylpropanoyl)piperidin-4-yl]phenyl}-2H-pyrrolo[3,4-c]pyridine-2-carboxamide, Nicotinamide phosphoribosyltransferase, SULFATE ION | | Authors: | Longenecker, K.L, Raich, D, Korepanova, A.V. | | Deposit date: | 2016-11-30 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and Characterization of Novel Nonsubstrate and Substrate NAMPT Inhibitors.
Mol. Cancer Ther., 16, 2017
|
|
4ILB
 
 | | The pentameric ligand-gated ion channel GLIC A237F in complex with Rubidium | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-12-29 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
3ESZ
 
 | | K2AK3A Flavodoxin from Anabaena | | Descriptor: | FLAVIN MONONUCLEOTIDE, FLAVODOXIN, GLYCEROL, ... | | Authors: | Herguedas, B, Martinez-Julvez, M, Hermoso, J, Goni, G, Medina, M. | | Deposit date: | 2008-10-06 | | Release date: | 2009-02-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Flavodoxin: A compromise between efficiency and versatility in the electron transfer from Photosystem I to Ferredoxin-NADP(+) reductase
Biochim.Biophys.Acta, 1787, 2009
|
|
1QOA
 
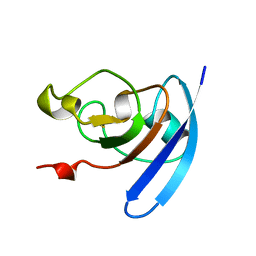 | | FERREDOXIN MUTATION C49S | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron-sulfur cluster cysteine-to-serine mutants of Anabaena -2Fe-2S- ferredoxin exhibit unexpected redox properties and are competent in electron transfer to ferredoxin:NADP+ reductase.
Biochemistry, 36, 1997
|
|
4HFI
 
 | | The GLIC pentameric Ligand-Gated Ion Channel at 2.4 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
3NSY
 
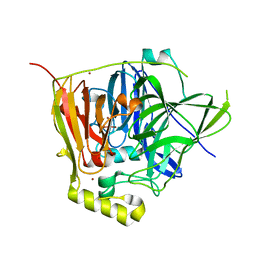 | | The multi-copper oxidase CueO with six Met to Ser mutations (M358S,M361S,M362S,M364S,M366S,M368S) | | Descriptor: | Blue copper oxidase cueO, COPPER (II) ION, CU-O-CU LINKAGE | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-02 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3NSF
 
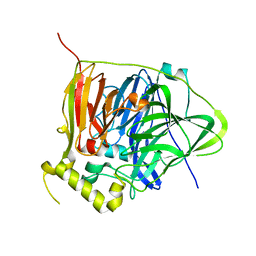 | | Apo form of the multicopper oxidase CueO | | Descriptor: | Blue copper oxidase cueO | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3NSC
 
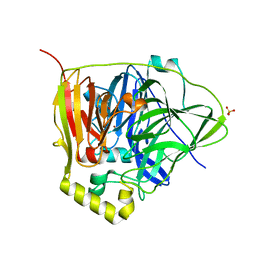 | | C500S MUTANT OF CueO BOUND TO Cu(II) | | Descriptor: | ACETATE ION, Blue copper oxidase cueO, COPPER (II) ION, ... | | Authors: | Roberts, S.A, Montfort, W.R, Singh, S.K. | | Deposit date: | 2010-07-01 | | Release date: | 2011-08-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
3NT0
 
 | |
3EPZ
 
 | | Structure of the replication foci-targeting sequence of human DNA cytosine methyltransferase DNMT1 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, GLYCEROL, SODIUM ION, ... | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Li, Y, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-09-30 | | Release date: | 2008-11-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | The replication focus targeting sequence (RFTS) domain is a DNA-competitive inhibitor of Dnmt1.
J.Biol.Chem., 286, 2011
|
|
3OD3
 
 | | CueO at 1.1 A resolution including residues in previously disordered region | | Descriptor: | 1,2-ETHANEDIOL, Blue copper oxidase cueO, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2010-08-10 | | Release date: | 2011-09-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Crystal structures of multicopper oxidase CueO bound to copper(I) and silver(I): functional role of a methionine-rich sequence.
J. Biol. Chem., 286, 2011
|
|
4LLV
 
 | | The structure of the unbound form of anti-HIV antibody 4E10 Fv | | Descriptor: | 4E10 Fv heavy chain, 4E10 Fv light chain, GLYCEROL, ... | | Authors: | Finton, K.A.K, Rupert, P.B, Strong, R.K. | | Deposit date: | 2013-07-09 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Autoreactivity and Exceptional CDR Plasticity (but Not Unusual Polyspecificity) Hinder Elicitation of the Anti-HIV Antibody 4E10.
Plos Pathog., 9, 2013
|
|
3QQX
 
 | | Reduced Native Intermediate of the Multicopper Oxidase CueO | | Descriptor: | Blue copper oxidase CueO, COPPER (I) ION, COPPER (II) ION, ... | | Authors: | Montfort, W.R, Roberts, S.A, Singh, S.K. | | Deposit date: | 2011-02-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | CueO E506D Mutant: Crystal Structure of Reduced Native Intermediate, Kinetics, and Impairment of Product Release
To be Published
|
|
