3MK5
 
 | | Crystal structure of 3,4-dihydroxy-2-butanone 4-phosphate synthase domain from Mycobacterium tuberculosis with sulfate and zinc at pH 4.00 | | Descriptor: | 3,4-dihydroxy-2-butanone 4-phosphate synthase, SULFATE ION, ZINC ION | | Authors: | Singh, M, Karthikeyan, S. | | Deposit date: | 2010-04-14 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural basis for pH dependent monomer-dimer transition of 3,4-dihydroxy 2-butanone-4-phosphate synthase domain from Mycobacterium tuberculosis
J.Struct.Biol., 174, 2011
|
|
4KGQ
 
 | | Crystal structure of a human light loop mutant in complex with dcr3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Tumor necrosis factor ligand superfamily member 14, ... | | Authors: | Liu, W, Zhan, C, Bonanno, J.B, Sampathkumar, P, Toro, R, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-04-29 | | Release date: | 2013-07-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Mechanistic basis for functional promiscuity in the TNF and TNF receptor superfamilies: structure of the LIGHT:DcR3 assembly.
Structure, 22, 2014
|
|
8Q93
 
 | | Crystal structure of the SARS-COV-2 RBD with neutralizing-VHHs Re30H02 and Re21D01 | | Descriptor: | Nanobody Re21D01, Nanobody Re30H02, Spike protein S1 | | Authors: | Aksu, M, Guttler, T, Rymarenko, O, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q95
 
 | | Crystal structure of the SARS-CoV-2 BA.1 RBD with neutralizing-VHHs Ma16B06 and Ma3F05 | | Descriptor: | Nanobody Ma16B06, Nanobody Ma3F05, Spike protein S1 | | Authors: | Aksu, M, Rymarenko, O, Guttler, T, Gorlich, D. | | Deposit date: | 2023-08-19 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Nanobodies to multiple spike variants and inhalation of nanobody-containing aerosols neutralize SARS-CoV-2 in cell culture and hamsters.
Antiviral Res., 221, 2023
|
|
8Q94
 
 | |
8QHP
 
 | |
8Q70
 
 | |
8P49
 
 | |
3B6S
 
 | | Crystal Structure of hla-b*2705 Complexed with the Citrullinated Vasoactive Intestinal Peptide Type 1 Receptor (vipr) Peptide (residues 400-408) | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, B-27 alpha chain, ... | | Authors: | Beltrami, A, Rossmann, M, Fiorillo, M.T, Sorrentino, R, Saenger, W, Ziegler, A, Uchanska-Ziegler, A. | | Deposit date: | 2007-10-29 | | Release date: | 2008-07-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Citrullination-dependent Differential Presentation of a Self-peptide by HLA-B27 Subtypes.
J.Biol.Chem., 283, 2008
|
|
4KF7
 
 | |
4KF8
 
 | |
5V2W
 
 | | Crystal structure of a LuxS from salmonella typhi | | Descriptor: | S-ribosylhomocysteine lyase, ZINC ION | | Authors: | Perumal, P, Raina, R, Manoj Kumar, P, Arockisamy, A, SundaraBaalaji, N. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a LuxS from salmonella typhi
To Be Published
|
|
7MKV
 
 | |
9IA8
 
 | | Crystal Structure of UFC1 K108R | | Descriptor: | Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Banerjee, S, Wiener, R. | | Deposit date: | 2025-02-08 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
9I9M
 
 | |
9I9O
 
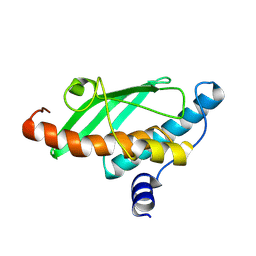 | | Crystal Structure of UFC1 K108M | | Descriptor: | Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Banerjee, S, Wiener, R. | | Deposit date: | 2025-02-06 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.051 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
9I9P
 
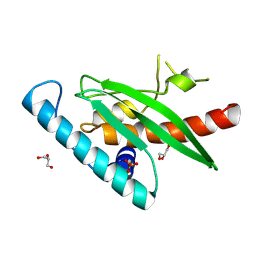 | | Crystal Structure of UFC1 W145H | | Descriptor: | GLYCEROL, SULFATE ION, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Banerjee, S, Weiner, R. | | Deposit date: | 2025-02-06 | | Release date: | 2025-05-07 | | Method: | X-RAY DIFFRACTION (2.022 Å) | | Cite: | UFC1 reveals the multifactorial and plastic nature of oxyanion holes in E2 conjugating enzymes.
Nat Commun, 16, 2025
|
|
9I9N
 
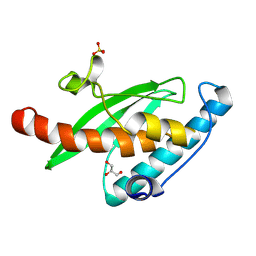 | |
9I1S
 
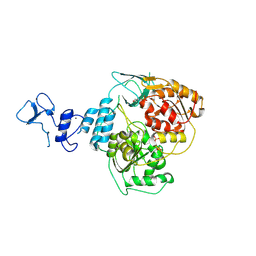 | | Crystal structure of the SARS-CoV-2 helicase NSP13 in complex with myricetin | | Descriptor: | 3,5,7-TRIHYDROXY-2-(3,4,5-TRIHYDROXYPHENYL)-4H-CHROMEN-4-ONE, 3[N-MORPHOLINO]PROPANE SULFONIC ACID, Helicase nsp13, ... | | Authors: | Kloskowski, P, Neumann, P, Ficner, R. | | Deposit date: | 2025-01-16 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Myricetin-bound crystal structure of the SARS-CoV-2 helicase NSP13 facilitates the discovery of novel natural inhibitors.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
9I4V
 
 | | Crystal structure of the SARS-CoV-2 helicase NSP13 | | Descriptor: | 3[N-MORPHOLINO]PROPANE SULFONIC ACID, PHOSPHATE ION, SARS-CoV-2 helicase NSP13, ... | | Authors: | Kloskowski, P, Neumann, P, Ficner, R. | | Deposit date: | 2025-01-27 | | Release date: | 2025-06-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Myricetin-bound crystal structure of the SARS-CoV-2 helicase NSP13 facilitates the discovery of novel natural inhibitors.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
5AFS
 
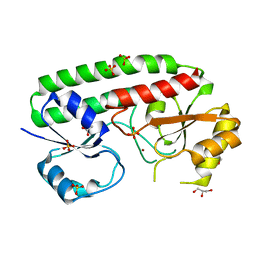 | | structure of Zn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, PERIPLASMIC SOLUTE BINDING PROTEIN, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis in Zn(2+)-bound state and biophysical characterization of CLas-ZnuA2.
Biochim. Biophys. Acta, 1864, 2016
|
|
2NBY
 
 | |
2NBX
 
 | |
2NBZ
 
 | |
2NC0
 
 | |
