1A1M
 
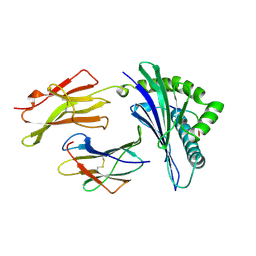 | | MHC CLASS I MOLECULE B*5301 COMPLEXED WITH PEPTIDE TPYDINQML FROM GAG PROTEIN OF HIV2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, BW-53 B*5301 alpha chain, ... | | Authors: | Smith, K.J, Reid, S.W, Harlos, K, Mcmichael, A.J, Stuart, D.I, Bell, J.I, Jones, E.Y. | | Deposit date: | 1997-12-11 | | Release date: | 1998-04-08 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Bound water structure and polymorphic amino acids act together to allow the binding of different peptides to MHC class I HLA-B53.
Immunity, 4, 1996
|
|
1AGC
 
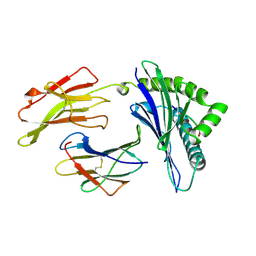 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYQL-7Q MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYQL - 7Q MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGF
 
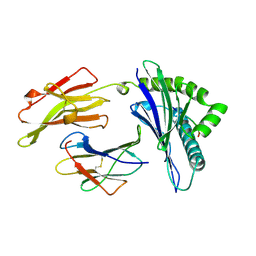 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKRYKL-5R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKRYKL - 5R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AKJ
 
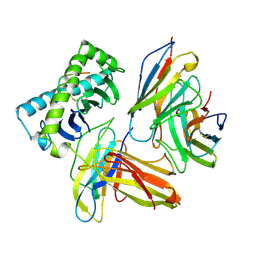 | | COMPLEX OF THE HUMAN MHC CLASS I GLYCOPROTEIN HLA-A2 AND THE T CELL CORECEPTOR CD8 | | Descriptor: | BETA 2-MICROGLOBULIN, HIV REVERSE TRANSCRIPTASE EPITOPE, MHC CLASS I HISTOCOMPATIBILITY ANTIGEN (HLA-A*0201) (ALPHA CHAIN), ... | | Authors: | Tormo, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-05-21 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of the complex between human CD8alpha(alpha) and HLA-A2.
Nature, 387, 1997
|
|
1AGD
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYKL-INDEX PEPTIDE) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYKL - INDEX PEPTIDE) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1AGE
 
 | | ANTAGONIST HIV-1 GAG PEPTIDES INDUCE STRUCTURAL CHANGES IN HLA B8-HIV-1 GAG PEPTIDE (GGKKKYRL-7R MUTATION) | | Descriptor: | B*0801, BETA-2 MICROGLOBULIN, HIV-1 GAG PEPTIDE (GGKKKYRL - 7R MUTATION) | | Authors: | Reid, S.W, Mcadam, S, Smith, K.J, Klenerman, P, O'Callaghan, C.A, Harlos, K, Jakobsen, B.K, Mcmichael, A.J, Bell, J, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1997-03-24 | | Release date: | 1997-06-16 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Antagonist HIV-1 Gag peptides induce structural changes in HLA B8.
J.Exp.Med., 184, 1996
|
|
1B6U
 
 | |
1B8K
 
 | | Neurotrophin-3 from Human | | Descriptor: | PROTEIN (NEUROTROPHIN-3) | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y, Choe, S. | | Deposit date: | 1999-02-01 | | Release date: | 1999-02-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structures of the neurotrophin 4 homodimer and the brain-derived neurotrophic factor/neurotrophin 4 heterodimer reveal a common Trk-binding site.
Protein Sci., 8, 1999
|
|
8QZR
 
 | | SARS-CoV-2 delta RBD complexed with BA.4/5-9 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BA.4/5-9 heavy chain, BA.4/5-9 light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-29 | | Release date: | 2024-04-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.77 Å) | | Cite: | Emerging variants develop total escape from potent monoclonal antibodies induced by BA.4/5 infection.
Nat Commun, 15, 2024
|
|
8R80
 
 | | SARS-CoV-2 Delta RBD in complex with XBB-9 Fab and an anti-Fab nanobody | | Descriptor: | Spike protein S1, XBB-9 Fab heavy chain, XBB-9 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-11-27 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (4.03 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRF
 
 | | SARS-CoV-2 delta RBD complexed with XBB-6 and beta-49 Fabs | | Descriptor: | Beta-49 heavy chain, Beta-49 light chain, Spike protein S1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-05-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QTD
 
 | | Local refinement of SARS-CoV-2 BA.2.86 Spike and XBB-7 Fab | | Descriptor: | Spike glycoprotein,Fibritin, XBB-7 fab heavy chain, XBB-7 fab light chain | | Authors: | Ren, J, Duyvesteyn, H.M.E, Stuart, D.I. | | Deposit date: | 2023-10-12 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QRG
 
 | | SARS-CoV-2 delta RBD complexed with XBB-2 Fab and NbC1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, NbC1, ... | | Authors: | Zhou, D, Ren, J, Stuart, D.I. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structure-function analysis shows SARS-CoV-2 BA.2.86 balances antibody escape and ACE2 affinity.
Cell Rep Med, 5, 2024
|
|
8QSQ
 
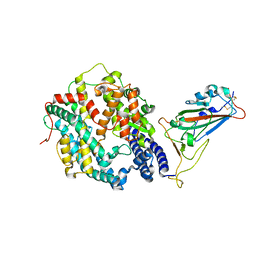 | |
1BND
 
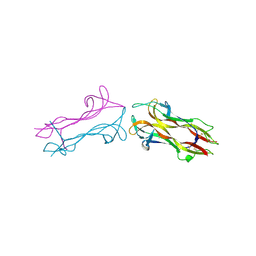 | | STRUCTURE OF THE BRAIN-DERIVED NEUROTROPHIC FACTOR(SLASH)NEUROTROPHIN 3 HETERODIMER | | Descriptor: | BRAIN DERIVED NEUROTROPHIC FACTOR, ISOPROPYL ALCOHOL, NEUROTROPHIN 3 | | Authors: | Robinson, R.C, Radziejewski, C, Stuart, D.I, Jones, E.Y. | | Deposit date: | 1994-12-12 | | Release date: | 1996-04-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the brain-derived neurotrophic factor/neurotrophin 3 heterodimer.
Biochemistry, 34, 1995
|
|
1UVJ
 
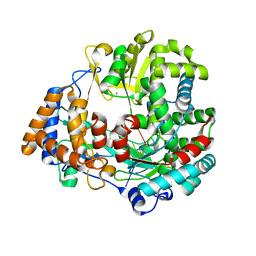 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 7nt RNA | | Descriptor: | 5'-R(*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UZJ
 
 | | Integrin binding cbEGF22-TB4-cbEGF33 fragment of human fibrillin-1, holo form. | | Descriptor: | CALCIUM ION, FIBRILLIN-1 | | Authors: | Lee, S.S.J, Knott, V, Harlos, K, Handford, P.A, Stuart, D.I. | | Deposit date: | 2004-03-12 | | Release date: | 2004-04-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the Integrin Binding Fragment from Fibrillin-1 Gives New Insights Into Microfibril Organization
Structure, 12, 2004
|
|
1W8X
 
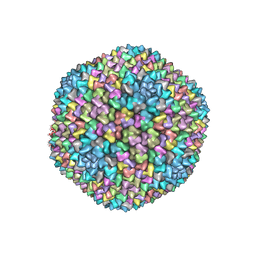 | | Structural analysis of PRD1 | | Descriptor: | MAJOR CAPSID PROTEIN (PROTEIN P3), PROTEIN P16, PROTEIN P30, ... | | Authors: | Abrescia, N.G.A, Cockburn, J.J.B, Grimes, J.M, Sutton, G.C, Diprose, J.M, Butcher, S.J, Fuller, S.D, San Martin, C, Burnett, R.M, Stuart, D.I, Bamford, D.H, Bamford, J.K.H. | | Deposit date: | 2004-10-01 | | Release date: | 2004-11-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Insights Into Assembly from Structural Analysis of Bacteriophage Prd1.
Nature, 432, 2004
|
|
1W4B
 
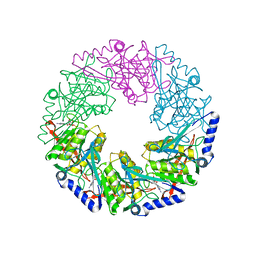 | | P4 protein from PHI12 in complex with product (AMPcPP Mg 22C) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W49
 
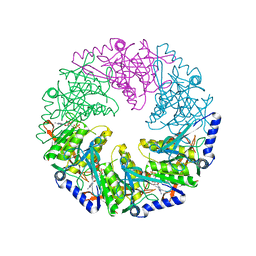 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP and Mg | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W4C
 
 | | P4 protein from Bacteriophage PHI12 apo state | | Descriptor: | NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W46
 
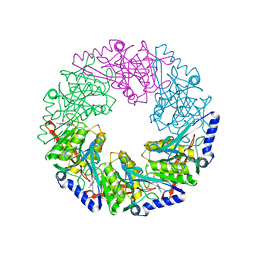 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MG | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W48
 
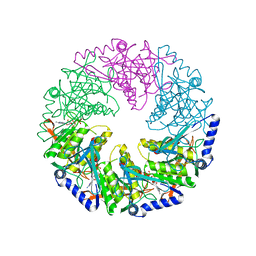 | | P4 protein from Bacteriophage PHI12 in complex with AMPcPP | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-10-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W47
 
 | | P4 protein from Bacteriophage PHI12 in complex with ADP and MN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MANGANESE (II) ION, NTPASE P4 | | Authors: | Mancini, E.J, Kainov, D.E, Grimes, J.M, Tuma, R, Bamford, D.H, Stuart, D.I. | | Deposit date: | 2004-07-22 | | Release date: | 2004-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Atomic Snapshots of an RNA Packaging Motor Reveal Conformational Changes Linking ATP Hydrolysis to RNA Translocation
Cell(Cambridge,Mass.), 118, 2004
|
|
1W9Z
 
 | | Structure of Bannavirus VP9 | | Descriptor: | VP9 | | Authors: | Jaafar, F.M, Attoui, H, Bahar, M.W, Siebold, C, Sutton, G, Mertens, P.P.C, Micco, P, Stuart, D.I, Grimes, J.M, Lamballerie, X. | | Deposit date: | 2004-10-21 | | Release date: | 2005-04-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | The Structure and Function of the Outer Coat Protein Vp9 of Banna Virus
Structure, 13, 2005
|
|
