3FUA
 
 | | L-FUCULOSE-1-PHOSPHATE ALDOLASE CRYSTAL FORM K | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, L-FUCULOSE-1-PHOSPHATE ALDOLASE, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Catalytic mechanism of the metal-dependent fuculose aldolase from Escherichia coli as derived from the structure.
J.Mol.Biol., 259, 1996
|
|
1FUA
 
 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T | | Descriptor: | BETA-MERCAPTOETHANOL, L-FUCULOSE-1-PHOSPHATE ALDOLASE, SULFATE ION, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
6AEC
 
 | | Binary complex of human DNA Polymerase Mu with MnATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA-directed DNA/RNA polymerase mu, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-04 | | Release date: | 2019-05-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AK8
 
 | | Pre-catalytic Ternary Complex of Human DNA Polymerase Mu with Templating Adenine and Incoming Ca-8oxodGTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AK9
 
 | | Pre-catalytic Ternary Complex of Human DNA Polymerase Mu with Templating Cytosine and Incoming Ca-8oxodGTP | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 8-OXO-2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AKH
 
 | | Pre-catalytic Ternary Complex of Human DNA Polymerase Mu with Templating Adenine and Incoming Mn-dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-31 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AEH
 
 | | Binary complex of human DNA Polymerase Mu with MnUTP | | Descriptor: | DNA-directed DNA/RNA polymerase mu, MANGANESE (II) ION, SULFATE ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-04 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
6AK6
 
 | | Binary Complex of Human DNA Polymerase Mu with MnCTP | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, DNA-directed DNA/RNA polymerase mu, MANGANESE (II) ION, ... | | Authors: | Chang, Y.K, Wu, W.J, Tsai, M.D. | | Deposit date: | 2018-08-30 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Human DNA Polymerase mu Can Use a Noncanonical Mechanism for Multiple Mn2+-Mediated Functions.
J.Am.Chem.Soc., 141, 2019
|
|
4WBQ
 
 | | Crystal structure of the exonuclease domain of QIP (QDE-2 interacting protein) solved by native-SAD phasing. | | Descriptor: | CALCIUM ION, QDE-2-interacting protein | | Authors: | Boland, A, Weinert, T, Weichenrieder, O, Wang, M. | | Deposit date: | 2014-09-03 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.693 Å) | | Cite: | Fast native-SAD phasing for routine macromolecular structure determination.
Nat.Methods, 12, 2015
|
|
1OXM
 
 | | STRUCTURE OF CUTINASE | | Descriptor: | BUTYL-PHOSPHINIC ACID 2,3-BIS-BUTYLCARBAMOYLOXY-PROPYL ESTER GROUP, CUTINASE | | Authors: | Longhi, S, Cambillau, C. | | Deposit date: | 1996-10-26 | | Release date: | 1997-05-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of cutinase covalently inhibited by a triglyceride analogue.
Protein Sci., 6, 1997
|
|
2FUA
 
 | | L-FUCULOSE 1-PHOSPHATE ALDOLASE CRYSTAL FORM T WITH COBALT | | Descriptor: | BETA-MERCAPTOETHANOL, COBALT (II) ION, L-FUCULOSE-1-PHOSPHATE ALDOLASE, ... | | Authors: | Dreyer, M.K, Schulz, G.E. | | Deposit date: | 1996-02-14 | | Release date: | 1996-10-14 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Refined high-resolution structure of the metal-ion dependent L-fuculose-1-phosphate aldolase (class II) from Escherichia coli.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
8GMN
 
 | | Crystal structure of human C1s in complex with inhibitor | | Descriptor: | Complement C1s subcomponent, SULFATE ION, [4-(1-aminophthalazin-6-yl)piperazin-1-yl](2-methylphenyl)methanone | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2023-03-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway.
J.Med.Chem., 66, 2023
|
|
6FN3
 
 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cryptochrome photoreceptor, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6FN0
 
 | | The animal-like Cryptochrome from Chlamydomonas reinhardtii in complex with 6-4 DNA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
6FN2
 
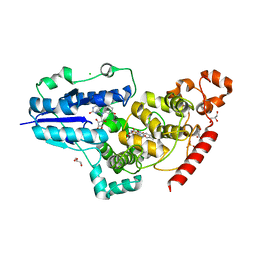 | | X-ray structure of animal-like Cryptochrome from Chlamydomonas reinhardtii | | Descriptor: | 8-HYDROXY-10-(D-RIBO-2,3,4,5-TETRAHYDROXYPENTYL)-5-DEAZAISOALLOXAZINE, CHLORIDE ION, Cryptochrome photoreceptor, ... | | Authors: | Franz, S, Ignatz, E, Wenzel, S, Zielosko, H, Yamamoto, J, Mittag, M, Essen, L.-O. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-01 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the bifunctional cryptochrome aCRY from Chlamydomonas reinhardtii.
Nucleic Acids Res., 46, 2018
|
|
1XZK
 
 | |
1C2N
 
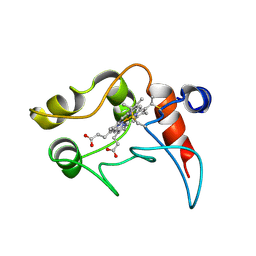 | | CYTOCHROME C2, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C2, HEME C | | Authors: | Cordier, F, Caffrey, M.S, Brutscher, B, Cusanovich, M.A, Marion, D, Blackledge, M. | | Deposit date: | 1998-04-27 | | Release date: | 1999-03-23 | | Last modified: | 2024-06-05 | | Method: | SOLUTION NMR | | Cite: | Solution structure, rotational diffusion anisotropy and local backbone dynamics of Rhodobacter capsulatus cytochrome c2.
J.Mol.Biol., 281, 1998
|
|
1QCE
 
 | |
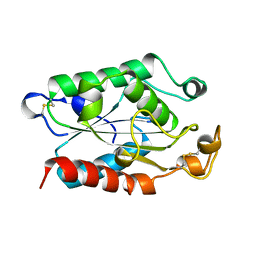
1FFA
 
 | |
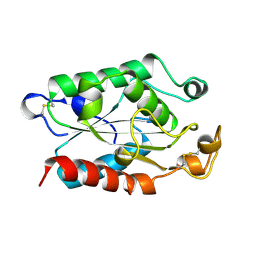
1FFC
 
 | |
1FFB
 
 | |
1FFE
 
 | |
1FFD
 
 | |
8EW1
 
 | | Structure of Bacple_01703-E145L | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Ulaganathan, T, Cygler, M. | | Deposit date: | 2022-10-21 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The porphyran degradation system of the human gut microbiota is complete, phylogenetically diverse and geographically structured across Asian populations
Biorxiv, 2023
|
|
7NFV
 
 | | Structure of SARS-CoV-2 Papain-like protease PLpro | | Descriptor: | CHLORIDE ION, GLYCEROL, PHOSPHATE ION, ... | | Authors: | Srinivasan, V, Gunther, S, Reinke, P, Werner, N, Falke, S, Brognaro, H, Ullah, N, Andaleeb, H, Perbandt, M, Alves Franca, B, Schwinzer, M, Wang, M, Sprenger, J, Lieske, J, Ginn, H, Lane, T.J, Yefanov, O, Gelisio, L, Koua, F, Saouane, S, Tolstikova, A, Groessler, M, Fleckenstein, H, Ewert, W, Trost, F, Lorenzen, K, Schubert, R, Han, H, Schmidt, C, Brings, L, Ehrt, C, Rarey, M, Galchenkova, M, Gevorkov, Y, Li, C, Perk, A, Awel, S, Hinrichs, W, Meents, A, Betzel, C. | | Deposit date: | 2021-02-07 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Antiviral activity of natural phenolic compounds in complex at an allosteric site of SARS-CoV-2 papain-like protease.
Commun Biol, 5, 2022
|
|
