1SQR
 
 | | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus. Northeast Structural Genomics Consortium Target PfR48. | | Descriptor: | 50S ribosomal protein L35Ae | | Authors: | Snyder, D.A, Aramini, J.M, Huang, Y.J, Xiao, R, Cort, J.R, Shastry, R, Ma, L.C, Liu, J, Rost, B, Acton, T.B, Kennedy, M.A, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-03-19 | | Release date: | 2004-11-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the 50S Ribosomal Protein L35AE from Pyrococcus furiosus: Northeast Structural Genomics Consortium Target PfR48
To be Published
|
|
1WWC
 
 | | NT3 BINDING DOMAIN OF HUMAN TRKC RECEPTOR | | Descriptor: | PROTEIN (NT-3 GROWTH FACTOR RECEPTOR TRKC) | | Authors: | Ultsch, M.H, Wiesmann, C, Simmons, L.C, Henrich, J, Yang, M, Reilly, D, Bass, S.H, De Vos, A.M. | | Deposit date: | 1999-04-30 | | Release date: | 1999-07-07 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of the neurotrophin-binding domain of TrkA, TrkB and TrkC.
J.Mol.Biol., 290, 1999
|
|
1IFQ
 
 | | Sec22b N-terminal domain | | Descriptor: | GLYCEROL, vesicle trafficking protein Sec22b | | Authors: | Gonzalez Jr, L.C, Weis, W.I, Scheller, R.H. | | Deposit date: | 2001-04-13 | | Release date: | 2001-05-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A novel snare N-terminal domain revealed by the crystal structure of Sec22b.
J.Biol.Chem., 276, 2001
|
|
1T84
 
 | | Solution structure of the Wiskott-Aldrich Syndrome Protein (WASP) autoinhibited core domain complexed with (S)-wiskostatin, a small molecule inhibitor | | Descriptor: | (2S)-1-(3,6-DIBROMO-9H-CARBAZOL-9-YL)-3-(DIMETHYLAMINO)PROPAN-2-OL, Wiskott-Aldrich syndrome protein | | Authors: | Peterson, J.R, Bickford, L.C, Morgan, D, Kim, A.S, Ouerfelli, O, Kirschner, M.W, Rosen, M.K. | | Deposit date: | 2004-05-11 | | Release date: | 2004-07-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Chemical inhibition of N-WASP by stabilization of a native autoinhibited conformation.
Nat.Struct.Mol.Biol., 11, 2004
|
|
2RII
 
 | |
1IFP
 
 | |
1XD8
 
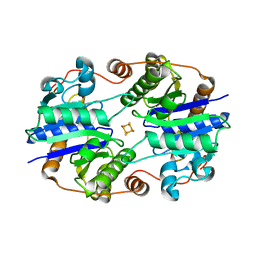 | | Crystal Structure of the Nitrogenase Fe protein Asp39Asn | | Descriptor: | IRON/SULFUR CLUSTER, Nitrogenase iron protein 1 | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
1QL1
 
 | |
1TDC
 
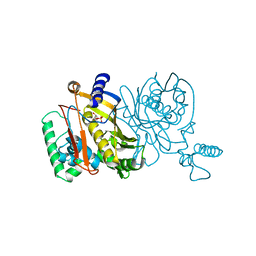 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | Descriptor: | 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, THYMIDYLATE SYNTHASE | | Authors: | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | Deposit date: | 1993-02-15 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
1TDA
 
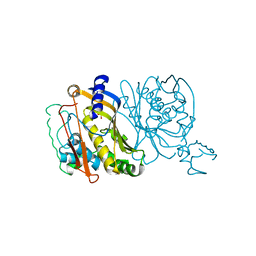 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | Descriptor: | PHOSPHATE ION, THYMIDYLATE SYNTHASE | | Authors: | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | Deposit date: | 1993-02-15 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
2TDD
 
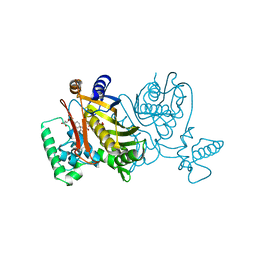 | | STRUCTURES OF THYMIDYLATE SYNTHASE WITH A C-TERMINAL DELETION: ROLE OF THE C-TERMINUS IN ALIGNMENT OF D/UMP AND CH2H4FOLATE | | Descriptor: | 5-FLUORO-2'-DEOXYURIDINE-5'-MONOPHOSPHATE, 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, THYMIDYLATE SYNTHASE | | Authors: | Perry, K.M, Carreras, C.W, Chang, L.C, Santi, D.V, Stroud, R.M. | | Deposit date: | 1993-04-05 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of thymidylate synthase with a C-terminal deletion: role of the C-terminus in alignment of 2'-deoxyuridine 5'-monophosphate and 5,10-methylenetetrahydrofolate.
Biochemistry, 32, 1993
|
|
1JTH
 
 | | Crystal structure and biophysical properties of a complex between the N-terminal region of SNAP25 and the SNARE region of syntaxin 1a | | Descriptor: | SNAP25, syntaxin 1a | | Authors: | Misura, K.M.S, Gonzalez Jr, L.C, May, A.P, Scheller, R.H, Weis, W.I. | | Deposit date: | 2001-08-21 | | Release date: | 2001-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure and biophysical properties of a complex between the N-terminal SNARE region of SNAP25 and syntaxin 1a.
J.Biol.Chem., 276, 2001
|
|
1VKJ
 
 | | Crystal structure of heparan sulfate 3-O-sulfotransferase isoform 1 in the presence of PAP | | Descriptor: | ADENOSINE-3'-5'-DIPHOSPHATE, SULFATE ION, heparan sulfate (glucosamine) 3-O-sulfotransferase 1 | | Authors: | Thorp, S, Lee, K.A, Negishi, M, Linhardt, R.J, Liu, J, Pedersen, L.C. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1
J.Biol.Chem., 279, 2004
|
|
1R7R
 
 | | The crystal structure of murine p97/VCP at 3.6A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Transitional endoplasmic reticulum ATPase | | Authors: | Huyton, T, Pye, V.E, Briggs, L.C, Flynn, T.C, Beuron, F, Kondo, H, Ma, J, Zhang, X, Freemont, P.S. | | Deposit date: | 2003-10-22 | | Release date: | 2003-12-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | The crystal structure of murine p97/VCP at 3.6A
J.Struct.Biol., 144, 2003
|
|
1RZT
 
 | | Crystal structure of DNA polymerase lambda complexed with a two nucleotide gap DNA molecule | | Descriptor: | 1,2-ETHANEDIOL, 5'-D(*CP*GP*GP*CP*AP*AP*CP*GP*CP*AP*C)-3', 5'-D(*GP*TP*GP*CP*G)-3', ... | | Authors: | Pedersen, L.C, Garcia-Diaz, M, Bebenek, K, Krahn, J.M, Blanco, L, Kunkel, T.A. | | Deposit date: | 2003-12-29 | | Release date: | 2004-03-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A structural solution for the DNA polymerase lambda-dependent repair of DNA gaps with minimal homology.
Mol.Cell, 13, 2004
|
|
1HY3
 
 | | CRYSTAL STRUCTURE OF HUMAN ESTROGEN SULFOTRANSFERASE V269E MUTANT IN THE PRESENCE OF PAPS | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, ESTROGEN SULFOTRANSFERASE | | Authors: | Pedersen, L.C, Petrochenko, E.V, Shevtsov, S, Negishi, M. | | Deposit date: | 2001-01-17 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction.
J.Biol.Chem., 277, 2002
|
|
1YEZ
 
 | | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30. | | Descriptor: | MM1357 | | Authors: | Rossi, P, Aramini, J.M, Swapna, G.V.T, Huang, Y.P, Xiao, R, Ho, C.K, Ma, L.C, Acton, T.B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-12-29 | | Release date: | 2005-02-22 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the conserved protein from the gene locus MM1357 of Methanosarcina mazei. Northeast Structural Genomics target MaR30.
To be Published
|
|
1JSV
 
 | |
1TP3
 
 | | PDZ3 domain of PSD-95 protein complexed with KKETPV peptide ligand | | Descriptor: | KKETPV peptide ligand, Presynaptic density protein 95 | | Authors: | Saro, D, Martin, P, Vickrey, J.F, Griffin, A, Kovari, L.C, Spaller, M.R. | | Deposit date: | 2004-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of the third PDZ domain of PSD-95 protein complexed with KKETPV peptide ligand
To be Published
|
|
1QCQ
 
 | | UBIQUITIN CONJUGATING ENZYME | | Descriptor: | PROTEIN (UBIQUITIN CONJUGATING ENZYME) | | Authors: | Cook, W.J, Jeffrey, L.C, Xu, Y, Chau, V. | | Deposit date: | 1999-05-10 | | Release date: | 1999-05-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tertiary structures of class I ubiquitin-conjugating enzymes are highly conserved: crystal structure of yeast Ubc4.
Biochemistry, 32, 1993
|
|
1XD9
 
 | | Crystal Structure of the Nitrogenase Fe protein Asp39Asn with MgADP bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, IRON/SULFUR CLUSTER, MAGNESIUM ION, ... | | Authors: | Jang, S.B, Jeong, M.S, Seefeldt, L.C, Peters, J.W. | | Deposit date: | 2004-09-05 | | Release date: | 2005-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and biochemical implications of single amino acid substitutions in the nucleotide-dependent switch regions of the nitrogenase Fe protein from Azotobacter vinelandii
J.Biol.Inorg.Chem., 9, 2004
|
|
1XW4
 
 | | Crystal Structure of Human Sulfiredoxin (Srx) in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Sulfiredoxin | | Authors: | Murray, M.S, Jonsson, T.J, Johnson, L.C, Poole, L.B, Lowther, W.T. | | Deposit date: | 2004-10-29 | | Release date: | 2005-05-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the retroreduction of inactivated peroxiredoxins by human sulfiredoxin.
Biochemistry, 44, 2005
|
|
1KXS
 
 | |
1YRS
 
 | | Crystal structure of KSP in complex with inhibitor 1 | | Descriptor: | 3-[(5S)-1-ACETYL-3-(2-CHLOROPHENYL)-4,5-DIHYDRO-1H-PYRAZOL-5-YL]PHENOL, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Cox, C.D, Breslin, M.J, Mariano, B.J, Coleman, P.J, Buser, C.A, Walsh, E.S, Hamilton, K, Kohl, N.E, Torrent, M, Yan, Y, Kuo, L.C, Hartman, G.D. | | Deposit date: | 2005-02-04 | | Release date: | 2005-04-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 1: The discovery of 3,5-diaryl-4,5-dihydropyrazoles as potent and selective inhibitors of the mitotic kinesin KSP
BIOORG.MED.CHEM.LETT., 15, 2005
|
|
1TBE
 
 | |
