2PCI
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | Probable diphthine synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Yamamoto, H, Matsuura, Y, Morikawa, Y, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-29 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2P6I
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Morikawa, Y, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-18 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2PCK
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Morikawa, Y, Matsuura, Y, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2P6K
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Ono, N, Nakamoto, T, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-18 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
2P6L
 
 | | Crystal structure of PH0725 from Pyrococcus horikoshii OT3 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, diphthine synthase | | Authors: | Yamamoto, H, Matsuura, Y, Ono, N, Shimada, H, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-19 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of PH0725 from Pyrococcus horikoshii OT3
To be Published
|
|
8GZV
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZX
 
 | | Escherichia coli FtsZ complexed with monobody (P212121) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZW
 
 | | Klebsiella pneumoniae FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
8GZY
 
 | | Escherichia coli FtsZ complexed with monobody (P21) | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Monobody | | Authors: | Matsumura, H, Yoshizawa, T, Fujita, J, Tanaka, S, Amesaka, H. | | Deposit date: | 2022-09-27 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
7BY0
 
 | | The cryo-EM structure of CENP-A nucleosome in complex with the phosphorylated CENP-C | | Descriptor: | DNA (145-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-21 | | Release date: | 2021-02-10 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
7BXT
 
 | | The cryo-EM structure of CENP-A nucleosome in complex with CENP-C peptide and CENP-N N-terminal domain | | Descriptor: | CENP-C, DNA (145-mer), Histone H2A type 1-B/E, ... | | Authors: | Ariyoshi, M, Makino, F, Fukagawa, T. | | Deposit date: | 2020-04-20 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the CENP-A nucleosome in complex with phosphorylated CENP-C.
Embo J., 40, 2021
|
|
5BRM
 
 | |
5BRK
 
 | | pMob1-Lats1 complex | | Descriptor: | MOB kinase activator 1A, Serine/threonine-protein kinase LATS1, ZINC ION | | Authors: | Ni, L, Luo, X. | | Deposit date: | 2015-05-31 | | Release date: | 2015-07-08 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for Mob1-dependent activation of the core Mst-Lats kinase cascade in Hippo signaling.
Genes Dev., 29, 2015
|
|
8SKQ
 
 | |
3LHM
 
 | |
2G92
 
 | |
1I1Z
 
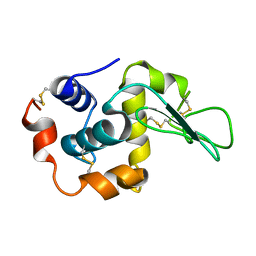 | | MUTANT HUMAN LYSOZYME (Q86D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I20
 
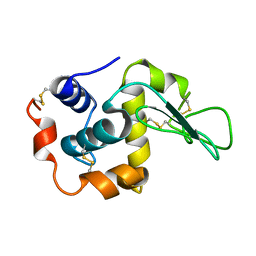 | | MUTANT HUMAN LYSOZYME (A92D) | | Descriptor: | LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
1I22
 
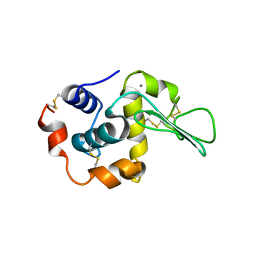 | | MUTANT HUMAN LYSOZYME (A83K/Q86D/A92D) | | Descriptor: | CALCIUM ION, LYSOZYME C | | Authors: | Kuroki, R. | | Deposit date: | 2001-02-05 | | Release date: | 2001-02-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and thermodynamic responses of mutations at a Ca2+ binding site engineered into human lysozyme.
J.Biol.Chem., 273, 1998
|
|
5DGR
 
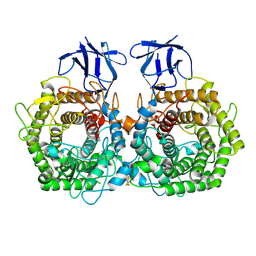 | | Crystal structure of GH9 exo-beta-D-glucosaminidase PBPRA0520, glucosamine complex | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, Putative endoglucanase-related protein, SODIUM ION | | Authors: | Suzuki, K, Honda, Y, Fushinobu, S. | | Deposit date: | 2015-08-28 | | Release date: | 2015-12-09 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of an inverting glycoside hydrolase family 9 exo-beta-D-glucosaminidase and the design of glycosynthase.
Biochem.J., 473, 2016
|
|
8H3M
 
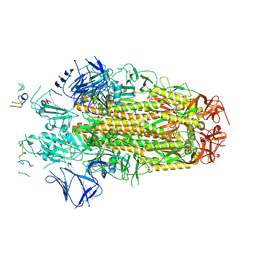 | | Conformation 1 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy chain, Spike glycoprotein | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
8H3N
 
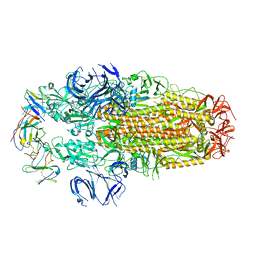 | | Conformation 2 of SARS-CoV-2 Omicron BA.1 Variant Spike protein complexed with MO1 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MO1 heavy-chain, MO1 light chain, ... | | Authors: | Ishimaru, H, Nishimura, M, Sutandhio, S, Shigematsu, H, Kato, K, Hasegawa, N, Mori, Y. | | Deposit date: | 2022-10-09 | | Release date: | 2023-05-10 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Identification and Analysis of Monoclonal Antibodies with Neutralizing Activity against Diverse SARS-CoV-2 Variants.
J.Virol., 97, 2023
|
|
7CJI
 
 | | Photosystem II structure in the S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7CJJ
 
 | | Photosystem II structure in the S2 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-07-11 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
7COU
 
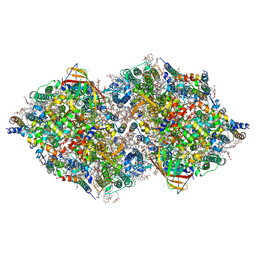 | | Structure of cyanobacterial photosystem II in the dark S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Li, H, Shen, J.-R, Suga, M. | | Deposit date: | 2020-08-05 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Capturing structural changes of the S 1 to S 2 transition of photosystem II using time-resolved serial femtosecond crystallography.
Iucrj, 8, 2021
|
|
