7TSV
 
 | | Room temperature rsEospa Trans-state structure at pH 5.5 | | Descriptor: | Trans-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
7TSR
 
 | | Room temperature rsEospa Cis-state structure at pH 8.4 | | Descriptor: | Cis-state rsEospa | | Authors: | Baxter, J.M, van Thor, J.J. | | Deposit date: | 2022-01-31 | | Release date: | 2022-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Observation of Cation Chromophore Photoisomerization of a Fluorescent Protein Using Millisecond Synchrotron Serial Crystallography and Infrared Vibrational and Visible Spectroscopy.
J.Phys.Chem.B, 126, 2022
|
|
1UVJ
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 7nt RNA | | Descriptor: | 5'-R(*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVK
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 dead-end complex | | Descriptor: | GUANOSINE-5'-MONOPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVM
 
 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5NT RNA conformation A | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVI
 
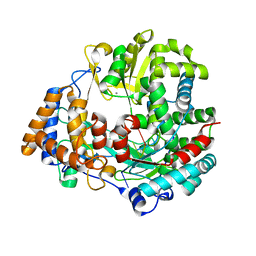 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 6nt RNA | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-19 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVN
 
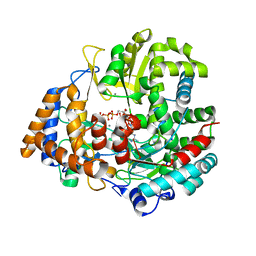 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 ca2+ inhibition complex | | Descriptor: | 5'-R(*UP*UP*UP*UP*CP*CP)-3', CALCIUM ION, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
1UVL
 
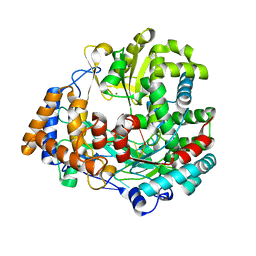 | | The structural basis for RNA specificity and Ca2 inhibition of an RNA-dependent RNA polymerase phi6p2 with 5nt RNA. Conformation B | | Descriptor: | 5'-R(*UP*UP*UP*CP*CP)-3', MANGANESE (II) ION, RNA-directed RNA polymerase | | Authors: | Salgado, P.S, Makeyev, E.V, Butcher, S, Bamford, D, Stuart, D.I, Grimes, J.M. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for RNA specificity and Ca2+ inhibition of an RNA-dependent RNA polymerase.
Structure, 12, 2004
|
|
7ADQ
 
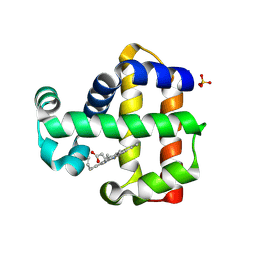 | | Serial Laue crystallography structure of dehaloperoxidase B from Amphitrite ornata | | Descriptor: | Dehaloperoxidase B, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Moreno-Chicano, T.M, Ebrahim, A.E, Srajer, V, Henning, R.W, Doak, B.C, Trebbin, M, Monteiro, D.C.F, Hough, M.A. | | Deposit date: | 2020-09-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Complementarity of neutron, XFEL and synchrotron crystallography for defining the structures of metalloenzymes at room temperature.
Iucrj, 9, 2022
|
|
8BCK
 
 | |
8BC5
 
 | |
8BBT
 
 | |
8BCL
 
 | |
6I43
 
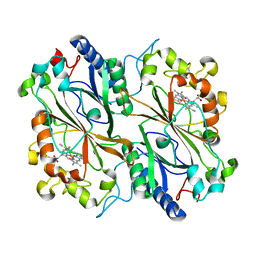 | | SFX Structure of Damage Free Ferric State of Dye Type Peroxidase Aa from Streptomyces lividans | | Descriptor: | Deferrochelatase/peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ebrahim, A, Moreno-Chicano, T, Chaplin, A.K, Sherrell, D.A, Duyvesteyn, H.M.E, Owada, S, Tono, K, Sugimoto, H, Strange, R.W, Worrall, J.A.R. | | Deposit date: | 2018-11-08 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins.
Iucrj, 6, 2019
|
|
7BH4
 
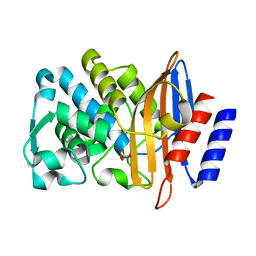 | | XFEL structure of apo CTX-M-15 after mixing for 0.7 sec with ertapenem using a piezoelectric injector (PolyPico) | | Descriptor: | Beta-lactamase, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHN
 
 | |
7BH5
 
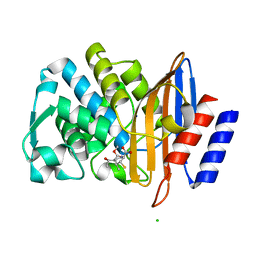 | | XFEL structure of the ertapenem-derived CTX-M-15 acylenzyme after mixing for 2 sec using a piezoelectric injector (PolyPico) | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHM
 
 | |
7BH7
 
 | | Room temperature, serial X-ray structure of the ertapenem-derived acylenzyme of CTX-M-15 (10 min soak) collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | (2~{S},3~{R},4~{R})-3-[5-[(3-carboxyphenyl)carbamoyl]pyrrolidin-3-yl]sulfanyl-4-methyl-5-[(2~{S},3~{R})-3-oxidanyl-1-oxidanylidene-butan-2-yl]-3,4-dihydro-2~{H}-pyrrole-2-carboxylic acid, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BH6
 
 | | Room temperature, serial X-ray structure of CTX-M-15 collected on fixed target chips at Diamond Light Source I24 | | Descriptor: | Beta-lactamase, CHLORIDE ION, SODIUM ION, ... | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHK
 
 | |
7BH3
 
 | | XFEL structure of CTX-M-15 resting state | | Descriptor: | Beta-lactamase, CHLORIDE ION, SULFATE ION | | Authors: | Hinchliffe, P, Tooke, C.L, Butryn, A, Spencer, J. | | Deposit date: | 2021-01-10 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An on-demand, drop-on-drop method for studying enzyme catalysis by serial crystallography.
Nat Commun, 12, 2021
|
|
7BHL
 
 | |
5MAS
 
 | | Peptaibol Bergofungin A | | Descriptor: | Bergofungin A | | Authors: | Gessmann, R, Petratos, K. | | Deposit date: | 2016-11-04 | | Release date: | 2017-02-22 | | Last modified: | 2017-03-01 | | Method: | X-RAY DIFFRACTION (0.84 Å) | | Cite: | A natural, single-residue substitution yields a less active peptaibiotic: the structure of bergofungin A at atomic resolution.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
3TZN
 
 | |
