1B4R
 
 | | PKD DOMAIN 1 FROM HUMAN POLYCYSTEIN-1 | | Descriptor: | PROTEIN (PKD1_HUMAN) | | Authors: | Bycroft, M. | | Deposit date: | 1998-12-28 | | Release date: | 1999-01-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of a PKD domain from polycystin-1: implications for polycystic kidney disease.
EMBO J., 18, 1999
|
|
1JLQ
 
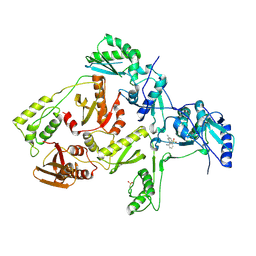 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH 739W94 | | Descriptor: | 2-AMINO-6-(3,5-DIMETHYLPHENYL)SULFONYLBENZONITRILE, HIV-1 RT, A-CHAIN, ... | | Authors: | Ren, J, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2001-07-16 | | Release date: | 2001-08-22 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 2-Amino-6-arylsulfonylbenzonitriles as non-nucleoside reverse transcriptase inhibitors of HIV-1.
J.Med.Chem., 44, 2001
|
|
3LUO
 
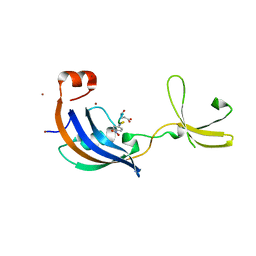 | | Crystal Structure and functional characterization of the thermophilic prolyl isomerase and chaperone SlyD | | Descriptor: | Peptidyl-prolyl cis-trans isomerase, Suc-Ala-Leu-Pro-Phe-pNA, ZINC ION | | Authors: | Loew, C, Neumann, P, Weininger, U, Stubbs, M.T, Balbach, J. | | Deposit date: | 2010-02-18 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure Determination and Functional Characterization of the Metallochaperone SlyD from Thermus thermophilus
J.Mol.Biol., 398, 2010
|
|
7SLV
 
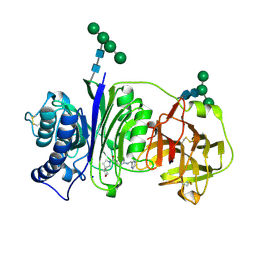 | | Vanin-1 complexed with Compound 3 | | Descriptor: | (8-oxa-2-azaspiro[4.5]decan-2-yl)(2-{[(pyrazin-2-yl)methyl]amino}pyrimidin-5-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
7SLY
 
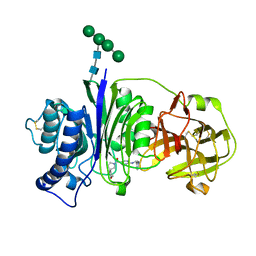 | | Vanin-1 complexed with Compound 27 | | Descriptor: | (8-oxa-2-azaspiro[4.5]decan-2-yl)(2-{[(1S)-1-(pyrazin-2-yl)ethyl]amino}pyrimidin-5-yl)methanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
7SLX
 
 | | Vanin-1 complexed with Compound 11 | | Descriptor: | (3R)-1-(2-{[1-(pyrimidin-5-yl)cyclopropyl]amino}pyrimidine-5-carbonyl)piperidine-3-carbonitrile, 2-acetamido-2-deoxy-beta-D-glucopyranose, Pantetheinase, ... | | Authors: | Vajdos, F.F. | | Deposit date: | 2021-10-25 | | Release date: | 2022-01-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Discovery of a Series of Pyrimidine Carboxamides as Inhibitors of Vanin-1.
J.Med.Chem., 65, 2022
|
|
1C3I
 
 | | HUMAN STROMELYSIN-1 CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, Kammlott, R.U, Crowther, R.L, Birktoft, J.J, Dunten, P, Engler, J.E. | | Deposit date: | 1999-07-27 | | Release date: | 2000-07-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
5EF9
 
 | | Structure of Influenza B Lee PB2 cap-binding domain | | Descriptor: | Polymerase basic protein 2 | | Authors: | Ma, X, Shia, S. | | Deposit date: | 2015-10-23 | | Release date: | 2015-11-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Basis of mRNA Cap Recognition by Influenza B Polymerase PB2 Subunit.
J.Biol.Chem., 291, 2016
|
|
8D14
 
 | | Helical ADP-Pi-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D18
 
 | | Straight ADP-F-actin 2 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
5EFA
 
 | |
8D15
 
 | | Bent ADP-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D13
 
 | | Helical ADP-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D16
 
 | | Bent ADP-Pi-F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.71 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
8D17
 
 | | Straight ADP-F-actin 1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Reynolds, M.J, Alushin, G.M. | | Deposit date: | 2022-05-26 | | Release date: | 2022-09-07 | | Last modified: | 2022-11-23 | | Method: | ELECTRON MICROSCOPY (3.69 Å) | | Cite: | Bending forces and nucleotide state jointly regulate F-actin structure.
Nature, 611, 2022
|
|
5EFC
 
 | |
8CUG
 
 | | Synthetic epi-Novo29 (2R,3S), synchrotron structure | | Descriptor: | ACETATE ION, Synthetic epi-Novo29 (2R,3S) | | Authors: | Kreutzer, A.G, Li, X, Krumberger, M, Nowick, J.S. | | Deposit date: | 2022-05-17 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.131 Å) | | Cite: | Synthesis and Stereochemical Determination of the Peptide Antibiotic Novo29.
J.Org.Chem., 88, 2023
|
|
8CUF
 
 | | Synthetic epi-Novo29 (2R,3S), X-ray diffractometer structure | | Descriptor: | ACETATE ION, IODIDE ION, Synthetic epi-Novo29 (2R,3S) | | Authors: | Kreutzer, A.G, Li, X, Krumberger, M, Nowick, J.S. | | Deposit date: | 2022-05-17 | | Release date: | 2023-01-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Synthesis and Stereochemical Determination of the Peptide Antibiotic Novo29.
J.Org.Chem., 88, 2023
|
|
1C8T
 
 | | HUMAN STROMELYSIN-1 (E202Q) CATALYTIC DOMAIN COMPLEXED WITH RO-26-2812 | | Descriptor: | 2-(2-{2-[(BIPHENYL-4-YLMETHYL)-AMINO]-3-MERCAPTO-PENTANOYLAMINO}-ACETYLAMINO)-3-METHYL-BUTYRIC ACID METHYL ESTER, CALCIUM ION, STROMELYSIN-1, ... | | Authors: | Steele, D.L, el-Kabbani, O, Dunten, P, Crowther, R.L. | | Deposit date: | 1999-07-29 | | Release date: | 2000-07-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Expression, characterization and structure determination of an active site mutant (Glu202-Gln) of mini-stromelysin-1.
Protein Eng., 13, 2000
|
|
1AMA
 
 | |
1BTV
 
 | | STRUCTURE OF BET V 1, NMR, 20 STRUCTURES | | Descriptor: | BET V 1 | | Authors: | Osmark, P, Poulsen, F.M, Gajhede, M, Larsen, J.N, Spangfort, M.D. | | Deposit date: | 1997-01-30 | | Release date: | 1997-08-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | X-ray and NMR structure of Bet v 1, the origin of birch pollen allergy.
Nat.Struct.Biol., 3, 1996
|
|
4M14
 
 | |
4M15
 
 | |
4M0Y
 
 | |
4M13
 
 | |
